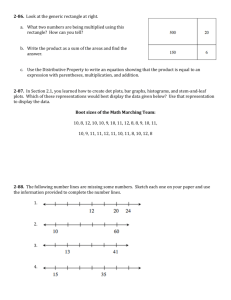
One goal of systems biology is to determine the biological
advertisement

Short abstract (50 words) We show the results of our effort to describe interactions among the network of proteins that participate in the intra-S checkpoint response to DNA damage, and the motif structure of the intra-S checkpoint proteome. We also demonstrate a new tool to visualize a hierarchical cluster tree of proteins. Long abstract (250 words) One goal of systems biology is to determine the biological significance of myriad interactions among cell proteins. High-through tools have used two-hybrid analysis to determine interactions among all of the S. cerevisiae open-reading frames. Similar efforts are underway to define protein interaction networks in humans and other vertebrate and invertebrate species. We show here the results of our effort to describe interactions among the network of proteins that participate in the intra-S checkpoint response to DNA damage. DNA damage checkpoint sense alterations in DNA structure and signal to the machinery of cell cycle to arrest or delay phase transitions. The intra-S checkpoint recognizes stalled DNA replication forks and sends signals to delay initiation of latefiring replicons and slow the rate of chain elongation by DNA polymerases. We have identified a set of 30 gene products that contribute to intra-S checkpoint response in human cells. Using the IDEA (idea.renci.org) website developed by RENCI and NCSA, we determined the yeast homologues of these human genes, created and visualized maps of the network of protein interactions in the human intra-S checkpoint using the opensource Cytoscape visualization tool. Protein interactions as demonstrated by two-hybrid analysis or co-immunoprecipitation are determined by specific domains of motifs that interact. The Motif-Network tool being created by RENCI and NCSA scientists enables researchers to determine the motifs that are represented in selected proteomes. Proteins that share motifs may share binding partners and biological functions. We show the motif structure of the intra-S checkpoint proteome.