emi412067-sup-0001-si

SUPPORTING MATERIALS
TABLES
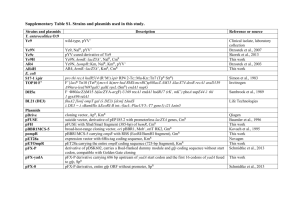
Bacterial strain
E. coli
DH5
P. putida
KT2442
KT2442::miniTn 7 BB-Gm
MRB1
MRB3
MRB3::TpMRB58
MRB3: miniTn 7 BB-Gm
MRB4
MRB7
MRB19
MRB21
MRB21::TpMRB53
MRB21::miniTn 7 BB Gm
MRB33
MRB33::TpMRB43
MRB33:: miniTn 7 BB Gm
MRB42
MRB43
Plasmid pENTR/D-TOPO pMRB43 pMRB53 pMRB58 pRK2013 pTNS2 pUC18Sfi-miniTn 7 -BBGm pUTminiTn 5 -Km
Genotype/phenotype
80d lacZ
∆
M15
∆
( lacZYA-argF )U169 recA 1 endA 1 hsdR 17 (r k
m k
+ ) supE 44 thi -1 gyrA relA 1 mt-2 hsdR 1 (r m + ) Rif r
KT2442 miniTn 7 BB Gm:: glmS Rif r Gm r
KT2442 miniTn 5 :: lapG Rif
KT2442 miniTn 5 :: dksA. Rif r Km r r Km r
KT2442 miniTn 5 :: dksA miniTn7BB-Gm[ dksA ]:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: dksA miniTn7BB-Gm:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: bifA Rif r Km r
KT2442 miniTn 5 :: dksA Rif r Km r
KT2442 miniTn 5 :: mvaB Rif r Km r
KT2442 miniTn 5 :: mvaB Rif r Km r
KT2442 miniTn 5 :: mvaB miniTn7BB-Gm[PP3539mvaB ]:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: mvaB miniTn7BB-Gm:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: bifA Rif r Km r
KT2442 miniTn 5 :: bifA miniTn7-BBGm[ bifA ]:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: bifA miniTn 7 BBGm:: glmS Rif r Km r Gm r
KT2442 miniTn 5 :: lapG Rif r Km r
KT2442 miniTn 5 :: lapG Rif r Km r
Genotype/phenotype
Vector for directional cloning. Km r pUC18Sfi-miniTn-BB 7 Gm-derived plasmid containing the bifA coding sequence and promoter region pUC18Sfi-miniTn-BB 7 Gm-derived plasmid containing the PP3539mvaB coding sequences and promoter region pUC18Sfi-miniTn-BB 7 Gm-derived plasmid containing the dksA coding sequence and promoter region
Helper plasmid for triparental mating. ColE1 replicon, Km R
R6K replicon-based helper plasmid, providing the Tn 7 transposition function in trans . Ap R , Mob+ pUC18Sfi-based delivery plasmid for the synthetic ransposon miniTn7-BBGm. Ap R , Gm R , Mob -
R6K-based suicide delivery plasmid for miniTn 5 -Km. Ap R Km R
Supporting Table S1. Bacterial strains and plasmids used in this work
Reference/source
Hanahan, 1983
Franklin et al, 1981
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
This work
Reference/source
Invitrogen
This work
This work
This work
Figurski and Helinski, 1979
Choi et al, 2005
D. Caballero and F.
Govantes, unpublished de Lorenzo et al, 1990
1
Oligonucleotide
CEKG-2A
CEKG-2B
CEKG-2C
CEKG-4 mTn5mapIr mTn5mapIIr
PP3538EcoRIFwd
PP3541SpeIFwd
PP4692RevXbaI
PP4693RevSpeI
Tn7-GlmS
Tn7R109
XS-0914
P-0914
Sequence (5’ to 3’)
Supporting Table S2. Oligonucleotides used in this work
GGCCACGCGTCGACTAGTACNNNNNNNNNNAGAG
GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC
GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT
GGCCACGCGTCGACTAGTAC
AGCTTGCTCAATCAATCAC
CACCTCAATCAATCACCGGATC
ATGTTCCGGAATTCCAACTGAACCAG
CCGCTGCCACTAGTTGGATGACGGCT
CAGGTGGATCTAGACGTGCCCGGCCG
AGGTGGAAACTAGTTCAGCCTTTGCC
AATCTGGCCAAGTCGGTGAC
CAGCATAACTGGACTGATTTCAG
CTTCTAGAGGCCTAGGCGGCCTCGGTTCCTTCGACAAGTTC
CAACTGCAGGCGAAAGATTTGCATTTGCT
Purpose
Mapping of miniTn 5 -Km insertions
Mapping of miniTn 5 -Km insertions
Mapping of miniTn 5 -Km insertions
Mapping of miniTn 5 -Km insertions
Mapping of miniTn 5 -Km insertions
Mapping of miniTn 5 -Km insertions
Cloning of PP3539mvaB
Cloning of PP3539mvaB
Cloning of dksA
Cloning of dksA
Testing miniTn 7 insertions
Testing miniTn 7 insertions
Cloning of bifA
Cloning of bifA
2
FIGURES
Supporting Figure S1. Phenotypic confirmation of the biofilm-persistent mutants.
Overnight cultures were diluted 50-fold in LB and inoculated in octuplicate in 96-well microtiter dishes. The dishes were incubated for 24 hours at 25ºC with moderate shaking
(150 rpm) and processed for planctonic (open bars) and biofilm (closed bars) biomass analysis. Bars represent the average and standard deviations of at least three biological replicates.
3
are needed to see this picture.
Supporting Figure S2. Location of the mini-Tn5-Km insertions in the biofilmpersistent mutants. Cartoons showing sections of the P. putida KT2440 chromosome around the transposon insertions are shown for PP0164 ( A ), PP0914 ( B ), PP3540 ( C ), and PP4693 ( D ). Genes bearing the insertions are shown as open pointed boxes. Other genes are shown as shaded pointed boxes. Absolute coordinates of the P. putida KT2440 genome are shown below the genes. Transposon insertions are denoted by inverted triangles. Each panel drawn to scale.
4
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Plasmids and bacterial strains used in this work and their relevant genotypes are summarized in Supporting Table 1. For general purposes, bacterial strains were routinely grown in LB medium (Sambrook et al., 2000) at 37°C (for E. coli ) or 30ºC for P. putida ).
When required, P. putida was grown in modified M9 minimal medium containing 25 mM sodium succinate as the sole carbon source and ammonium chloride (1 g l -1 ) as the sole nitrogen source (Mandelbaum et al., 1993). For solid media, Bacto-Agar (Difco) was added to a final concentration of 18 g l -1 . Antibiotics and other additions were used, when required, at the following concentrations: ampicillin (100 mg l -1 ), kanamycin (20 mg l -1 ), rifampicin (10 mg l -1 ), gentamycin (10 mg l -1 ), and 5-bromo-4-chloro-3-indoyl-
β-Dgalactopyranoside (X-gal) (25 mg l-1). All reagents were purchased from Sigma-Aldrich.
Plasmid and strain construction
The synthetic transposon miniTn 7 BB-Gm was assembled as part of the UPO-
Sevilla team project for the iGEM 2011 competition (David Caballero and Fernando
Govantes, unpublished)(further details on this construct are available at http://partsregistry.org/Part:BBa_K510000). Plasmids pMRB43, pMRB53 and pMRB58 were constructed by amplifying DNA fragments containing bifA, PP3539mvaB or dksA and their corresponding promoter regions (see Supporting Table 2 for primers) and cloning in the miniTn 7 BB-Gm delivery vector pUC18Sfi-miniTn 7 BB-Gm. MiniTn 7 -bearing P. putida strains were constructed by electroporation as previously described (Choi et al, 2006).
Identification of the miniTn5-Km insertion points
Mini-Tn 5 insertions were mapped by a modified arbitrary primed PCR approach
(O’Toole et al, 1999). In order to enrich in fragments harboring the transposon ends, oligonucleotide mapTn5Ir, annealing to both ends of miniTn 5 Km, was used on its own for the initial ten PCR cycles. Subsequently, a mix of primers CEKG-2A, CEKG-2B and
5
CEKG-2C was added to the reaction and subjected to six PCR cycles with annealing temperature starting at 42ºC and decreasing 1ºC each cycle. Finally, 30 additional PCR cycles were performed with an annealing temperature of 56ºC. Products flanking the transposon were specifically reamplified with oligonucleotides mapTn5IIr and CEKG-4.
The CACC tag at the 5' end of mapTn5IIr enabled directional cloning into the pENTR TM /D-
TOPO
®
vector (Invitrogen). The cloned fragments were subjected to commercial sequencing using mapTn5IIr as the primer (Sistemas Genómicos, Valencia, Spain).
6
BIBLIOGRAPHY
Choi, K.-H., Gaynor, J.B., White, K.G., Lopez, C., Bosio, C.M., Karkhoff-Schweizer, R.R. and Schweizer, H.P. (2005) A Tn 7 -based broad-range bacterial cloning and expression system. Nature Methods 2: 443-448.
Choi, K.H., Kumar, A. and Schweizer, H.P. (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64: 391-
397. de Lorenzo, V., Herrero, M., Jakubzik, U. and Timmis, K. N. (1990) Mini-Tn 5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J Bacteriol 172: 6568-6572.
Figurski, D.H. and Helinski, D.R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc Natl Acad Sci USA
76: 1648-1652.
Franklin, F.C., Bagdasarian, M., Bagdasarian, M.M. and Timmis, K.N. (1981) Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci
USA 78: 7458-7462.
Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol
166: 557-580.
Mandelbaum, R.T., Wackett L.P. and Allan, D.L. (1993) Mineralization of the s -triazine ring of atrazine by stable bacterial mixed cultures. Appl Environ Microbiol 59: 1695-1701.
O’Toole, G.A., Pratt, L.A., Watnick, P.I., Newman, D.K., Weaver, V.B., and Kolter, R.
(1999) Genetic approaches to study of biofilms. Methods Enzymol 310: 91-109.
Sambrook, J., Russell, D.W. and Russell, D. (2000) Molecular cloning, a laboratory
7
manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press.
8