phenotypes loci

42 PNL Volume 20 1988 RESEARCH REPORTS
LINKAGE BETWEEN Tpi-p AND Le
Weeden, N. F. and D. M. Hagens NYS Agricultural Experiment Station
Geneva, NY 14456 USA
Two triosephosphate isomerase (TPI) isozymes are present in pea leaf e x t r a c t s (1). The i s o z y m e s c a n be c l e a r l y r e s o l v e d by electrophoresis on h o r i z o n t a l starch gels using the buffer system of Selander et al. (2). The isozymes were v i s u a l i z e d by the a s s a y described previously ( 1 ) . We i d e n t i f i e d an electrophoretic variant (designated "a") for the more anodal, plastidspecific isozyme in the line JI 1794 ( k i n d l y provided by P. Matthews, John Innes I n s t i t u t e ) . Hybrids between JI 1794 and several o t h e r l i n e s ( e a c h f i x e d for the slower migrating variant " b " ) d i s p l a y e d a 3-banded "heterozygous" phenotype expected for this dimeric enzyme ( F i g . 1). JI 1794 was used as the maternal parent in some of t h e c r o s s e s and as the paternal parent in others. The appearance of the heterozygous phenotype in all hybrids provided evidence that this plastid-specific isozyme was coded by a nuclear gene, Tpi-p. Further evidence for the n u c l e a r l o c a t i o n of the coding gene was generated by the Mendelian segregation ratios obtained for the TPI phenotypes in several
F2 p o p u l a t i o n s (Table 1).
Two of the F2 progenies were segregating for a considerable number of known marker loci. The JI
1794 x s l o w segregated at Aat-p , Idh , I, Gal-2 , Amy-1 , Aat-m, Skdh, Oh, S, Wb, Fum, M, Acp-3, Gty,
Le, Acp-1, Pgd-c, Pl, Pgd-p, Pep-3, R, and B t . The JI 1 7 9 4 x A 7 8 3 - 1 6 I progeny segregated for many of the above markers plus Arg, Sil, Pal, and Lap-1. Significant deviation from random assortment was observed between Tpi-p and Le in both of t h e F2 progenies ( T a b l e 2 ) . None of the o t h e r markers gave a recombination frequency with Tpi-p of less than 30%.
These results indicate that Tpi-p is located near Le on chromosome 4. The calculated map distance differed c o n s i d e r a b l y b e t w e e n the two crosses, so that we a r e not a b l e to g i v e a p r e c i s e e s t i m a t e of this distance. Nor were we able to d e t e r m i n e the p o s i t i o n of Tpi-p r e l a t i v e to other markers on chromosome 4. Despite these u n c e r t a i n t i e s , we feel that these linkages are not caused by karyotype d i f f e r e n c e s between the parents because all other markers displayed normal segregation r a t i o s and the fertility in the F2 plants a p p e a r e d n o r m a l . The p o s i t i o n of Tpi-p is far from other isozyme l o c i , t h e nearest b e i n g an esterase locus described by Hunt and Barnes ( 3 ) .
O t h e r loci c o d i n g plastid-s p e c i f i c proteins have been mapped to chromosomes 1, 2, 5, and 7, but none have been previously placed on chromosome 4. Once we have t r a n s ferred the rare a allele to a dwarf background, it should prove to be an excellent marker because the phenotypes are e a s i l y d i s t i n g u i s h e d and t h e r e are few other convenient marker l o c i on that r e g i o n of the chromosome.
1.
Weeden, N.F. and L. D . Gottlieb. 1980. Plant Physiol. 66:400-403.
2.
Selander, R.K., M.H. Smith, S . Y . Yang, W . E . Johnson, and J.B. Gentry. 1971. Studies in Genetics
V I . Univ. Texas Pub. 7 103:49-90.
3.
Hunt, J.S. and M.F. Barnes. 1982. Euphytica 31:341-348.
PNL Volume 20 1988 RESEARCH REPORTS 43
Table 1. Segregation of triosephosphate isomerase phenotypes
Cross TPI phenotype*
2 X
(1:2:1)
JI 2018 x JI 1794
JI 1794 x slow
JI 1794 x A783-161 a
9
19
11 ab
19
32
26 b
9
12
16
0.02 1
.57
0.96
Designations: a = faster migrating variant, ab = 3-banded phenotype, b = slower migrating variant.
Table 2. Joint segregation of Le and Tpi-p.
Cross N No. plants with designated phenotype*
JI 1794 x slow
JI 1794 x 161
63
53
+ /aa +/ab + /bb -/aa
19
11
32
23
0
5
0
0
-/ab
0
3
-/bb
12
11 x
2 Recomb
Fract.
63.0
21.7
1 +/- 1
15 +/-
5
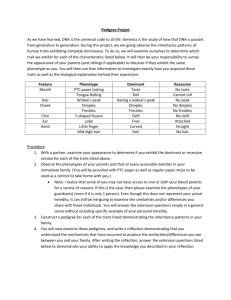
Fig. 1. Segregation of triosephosphate isomerase phenotypes in the
JI 1794 x slow progeny. The parental phenotypes are shown on the left side of the gel. Anode is toward top of figure,
*****