Supplementary Information (doc 556K)
advertisement

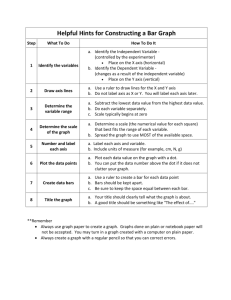
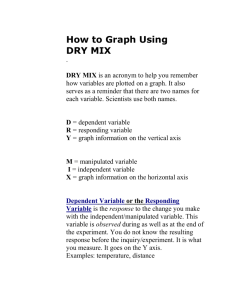
SUPPLEMENTARY INFORMATION Interpolation of map: Minimum curvature interpolation was applied separately for each ordination axis, which resulted from the three interpolated grid layers of ordination axis scores. Minimum Curvature interpolation using TNT Mips v.6.9 was applied as follows: first, a grid with pixel size 20 x 20 km was constructed from sampling point values (axis scores in our case) using the inverse distance, weighting power of 2, search distance of 100 cells and coarse grid ratio of two. In other words, the algorithm calculated values for every second grid intersection as a weighted mean of the sampling points located within the distance of 100 grid cells. Secondly, the initial grid was smoothed using 100 iterations, a matching tolerance (the difference between new values and original values in an iteration run) of 0.1, and no variation between original values and iterative results in sampling points. Interpretation of the map: The produced map may be interpreted with the help of a RGB color cube (Supplementary Figure S1), where three axes of the colour cube (red, green and blue) correspond to the three ordination axes in such a way that scores from axis 1 are shown in red, scores from axis 2 in green and scores from axis 3 in blue. Intensities of the colours increase when axis scores become higher. Blending of the main colours corresponds to the three-dimensional value of axis scores (Supplementary Figure S2). For instance the red areas in the map have high values in axis 1 (breed FFR in Fig. S2), correspondingly green areas have high values in axis 2 (breed RDM in Fig. S2) and blue areas have high values in axis 3 (breed SLB in Fig. S2). The areas having magenta (purple) colour in the map have high axis scores values in axis 1 and 3 but not in axis 2, and yellow areas in turn have high values in axis 1 and 2 but not in axis 3. The light areas have higher axis values in general than darker areas (breeds ESN and REG in Fig. S2), and the black colour occurs when all axes values are close to zero, and the white colour when axes score values are close to the maximum value of 255. The most similar breeds in terms of genetic data are shown in similar colours, whereas the genetically most distinct breeds are shown in colours located in the opposite corners or sides of the colour cube. SUPPLEMENTARY FIGURE S1 Figure S1. RGB color cube for help of map interpretation. Scores from PCO axis 1 are shown in red, scores from PCO axis 2 in green and scores from PCO axis 3 in blue. The sampling points, with the population acronyms, are shown as for map 4A in the main text. SUPPLEMENTARY FIGURE S2 Figure S2. Illustration of blending of shades in RGB map for help of map interpretation. Legend in the separate box tells that each figure tells separately the scores of PCA axis 1, axis 2 and 3 and the colours corresponding to the scores for selected breeds. The circle in the middle shows the colour in RGB map 4A corresponding to three axes scores and abbreviation of the breed. Note that ranges of the axes scores varied; range of axis 1 [-0.217, 0.119], axis 2 [-0.112, 0.228] and axis 3 [-0.103, 0.155]. SUPPLEMENTARY FIGURE S3 Figure S3. A neighbor-joining tree of mtDNA haplotypes. Only the most common T3 haplotypes (frequency ≥ 5) were included in the analysis (see also the Figures 1 and 2). The numbers above the branches indicate bootstrap proportions (only values > 50% shown) in 1,000 replications. SUPPLEMENTARY FIGURE S4 12000 Frequency 10000 8000 6000 4000 2000 31 29 27 25 23 21 19 17 15 13 11 9 7 5 3 1 0 Pairwise differences Figure S4. The mismatch distribution of mtDNA sequences in 235 animals analysed. A bimodal distribution indicates the deep genetic divergence between the taurine and zebu sequences. Unweighted mean pairwise difference between the sequences was 2.641. SUPPLEMENTARY FIGURE S5 Frequency The entire data set of 405 chromosomes 20000 18000 16000 14000 12000 10000 8000 6000 4000 2000 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Sum of size differences Figure S5. The mismatch distribution of the entire Y-data set. SUPPLEMENTARY TABLE S1 Table S1. Y-chromosomal and mtDNA diversity estimates for the breeds and their geographic origins. Y-chromosomal haplotypes Population EUROPEAN BREEDS Byelorussian Red Danish Red Danish Jersey Jutland breed (Denmark) Estonian Red Estonian Holstein-Friesian Estonian Native Faroe Islands Cattle Finnish Ayrshire Finnish Holstein-Friesian Eastern Finncattle Northern Finncattle Western Finncattle German Simmental Icelandic Cattle Latvian Brown Latvian Danish Red Latvian Blue Lithuanian Red Lithuanian Black-and-White Lithuanian Light Grey Sample size 22 15 12 6 10 NA 10 3 14 11 11 3 9 3 11 8 10 9 9 10 10 Number of haplotypes MtDNA Haplotypic diversity Total Private 7 3 3 3 3 0 0 0 0 0 0.909 0.378 0.682 0.733 0.378 3 3 3 4 4 2 3 3 3 2 5 2 7 4 4 0 0 0 0 2 0 0 0 1 0 1 0 1 0 0 0.378 0.385 0.709 0.600 0.556 0.655 0.250 0.800 0.556 0.909 0.733 0.733 Sample size 8 9 NA 9 5 1 4 14 7 7 18 8 7 NA NA NA NA NA NA NA NA Number of haplotypes Haplotypic diversity Mean number of differences between haplotypes Nucleotide diversity x 10-3 Total Private 5 2 2 0 0.786 0.500 1.250 0.500 4.92 1.97 6 4 1 1 3 3 2 2 5 4 2 1 1 0 1 1 1 0 0 1 0.833 0.900 1.500 2.800 5.94 11.02 0.000 0.385 0.524 0.476 0.209 0.786 0.810 0.000 0.956 1.714 0.952 0.418 1.179 1.905 0.00 3.76 6.75 3.75 1.65 4.64 7.50 Lithuanian White Backed Norwegian Red Cattle (NRF) Western Red Polled (Norway) Eastern Red Polled (Norway) Doela Cattle (Norway) Troender Cattle (Norway) Telemark (Norway) Western Fjord Cattle (Norway) Polish Holstein-Friesian Suksun (Russia) Istoben (Russia) Pechora type (Russia) Kholmogory (Russia) Yaroslavl (Russia) Red Gorbatov (Russia) Yurino (Russia) Yakutian Cattle (Russia) Busa (Serbia) Podolian (Serbia) Swedish Holstein-Friesian Swedish Red-and-White Vane Cattle (Sweden) Ringamala Cattle (Sweden) Swedish Red Polled Swedish Mountain Cattle Bohus Poll (Sweden) Fjall Cattle (Sweden) Ukrainian Whitehead Ukrainian Grey 10 12 4 6 4 7 2 6 10 5 9 7 8 4 2 6 17 5 4 NA NA NA NA 5 9 NA 4 11 7 3 3 2 3 2 2 1 4 4 2 3 2 3 4 1 1 3 2 2 0 0 0 1 1 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0.644 0.621 3 2 0 0 0.800 0.286 2 7 4 0 3 4 0.909 0.810 CENTRAL ASIAN BREEDS Ala-Tau (Kazakshtan) Bushuev (Uzbekistan) 6 4 3 3 0 1 0.600 0.286 0.867 0.533 0.400 0.667 0.571 0.607 0.600 0.600 0.600 NA NA NA NA NA NA NA NA NA 10 9 9 14 10 2 4 24 8 11 5 5 5 7 5 4 6 6 10 8 8 6 3 4 2 2 3 7 2 4 3 2 2 2 3 3 1 5 7 1 4 3 1 2 1 1 0 5 1 3 1 0 2 0 0 2 0 4 4 0 0.956 0.833 0.556 0.626 0.356 2.622 1.944 0.833 0.868 0.711 10.32 7.66 3.28 3.42 2.80 0.645 0.536 0.709 0.800 0.600 0.400 0.286 0.700 0.833 0.000 0.933 0.933 0.000 1.630 1.071 1.091 2.400 1.200 2.000 0.571 0.800 1.000 0.000 2.200 10.911 0.000 6.42 4.22 4.29 9.45 4.72 7.87 2.25 3.15 3.94 0.00 8.66 42.96 0.00 5 4 3 3 2 0 0.800 0.814 14.800 1.613 58.27 6.36 NEAR-EASTERN BREEDS Anatolian Black (Turkey) East Anaolian Red (Turkey) South Anatolian Red (Turkey) Turkish Grey Damascus (Syria) Middle Iraqi North Iraqi South Iraqi 6 4 4 4 5 5 3 4 6 4 4 4 5 5 3 4 4 0 1 1 2 0 3 2 1.000 1.000 1.000 NA NA NA NA NA NA NA NA NA= Not analysed in this study The frequencies of mtDNA and Y-chromosomal haplotypes in each breed can be obtained on request from the corresponding author. SUPPLEMENTARY TABLE S2 Table S2. Allele frequencies of the Y-chromosomal microsatellites in the present cattle data set. Locus INRA124 Allele size (bp) 130 132 Frequency 0.025 0.975 INRA189 82 88 90 98 100 102 104 106 0.015 0.022 0.017 0.672 0.027 0.104 0.119 0.025 BM861 156 158 160 0.020 0.978 0.002 DYZ1 360 362 364 366 0.133 0.170 0.002 0.694 BYM-1 252 254 256 258 0.010 0.035 0.178 0.778