Objectives 15
advertisement

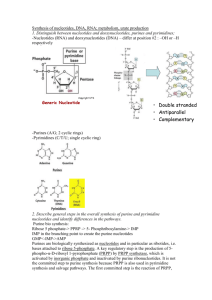
Purine metabolism synthase- no high energy compound involved synthetase – uses high energy compound Purine biosynthesis - Ribose-5-P (from pentose phosphate pathway) PRPP via PRPP synthetase 5-P-betaribosylamine via PRPP amidotransferase - PRPP used in synthesis of purine and pyrimidine nucleotides by providing ribose sugar and alpha phosphate - PRPP amidotransferase is rate determining and committed step of purine synthesis - remaining reaction require ATP, amino acids (gly, glu, asp), and folic acid-derived cofactor (N10- formyltetrahydrofolate, formyl THF) - 6 high energy P bonds cleaved in production of inosinic acid (IMP) first purine nucleotide (base, sugar, P) formed - AMP and GMP from IMP (separate pathways), figure 2 on 15-3 - GMP synthesis required redox reaction and nitrogen incorporation from amide side chain of glutamine - AMP acquires nitrogen from aspartate with production of fumarate; aspartate regenerated from fumarate by formation, in citric acid cycle, of oxaloacetate, which is then aminated to aspartate (fumarate malate oxaloacetate aspartate) - mono-P products phosphorylated to di- and triphosphates by nucleoside monophosphate kinase and nucleoside diphosphate kinase - end result is production of purine ribonucleotides - purine deoxyribionucleotides catalyzed by ribonucleotide reductase 1. a. Regulation of purine synthesis - adenine and guanine nucleotides both inhibit the synthesis of IMP early in pathway at both PRPP synthetase and PRPP amidotransferase; they also feedback inhibit their own product from IMP - excess ATP activates pathway from IMP GMP, while excess GTP activates pathway from IMP AMP to maintain proper balance of purine nucleotides b. Purine degradation - starts with removal of phosphate from purine nucleotide forms, yielding nucleoside (base, sugar) - adenosine inosine catalyzed by adenosine deaminase - sugars removed by purine nucleoside phosphorylase purine bases guanine and hypoxanthine, and ribose-1-P - base products xanthine oxidized by xanthine oxidase uric acid; xanthine oxidase also converts hypoxanthine xanthine; xanthine oxidase requires molecular oxygen and molybdenum, non-heme iron, and FAD Uric acid secretion - uric acid is the purine degradative product weak acid, with pK of 5.8; ionized form more water-soluble than protonated form - urine at pH 4.8 uric acid is 90% protonated can dissolve 1/10 as much urate as urine at pH 6.8 (90% ionized) - normal urine pH below 5.8 overproduction of uric acid can lead to formation of stones (not as soluble) c. Purine salvage pathway - besides de novo synthesis, purine nucleotides can be formed directly from purine bases via salvage pathway - two enzymes: hypoxanthine-guanine phosphoribosyl transferase (HGPRT) and adenine phosphoribosyl transferase - HGPRT adds PRPP (from PRPP synthetase) to hypoxanthine IMP or to guanine GMP - prevents irreversible destruction of hypoxanthine, guanine and adenine (purine bases reutilized) - salvage pathway saves energy because of high energy demand of de novo synthesis pathway - important for salvage of dietary nucleotides; salvage purine bases uric acid kept low (prevents gout) - HGPRT defect very low activity; Lesch-Nyhan syndrome d. Disorders associated with defects of enzymes in purine metabolic pathways: - elevation of uric acid from overproduction of purines or decreased excretion of uric acid - hyperuricemia associated with type I glycogen storage disease (von Gierke’s); glucose-6Pase defect increased oxidation of glucose-6-P ribose-5-P elevates purine production through saturation of PRPP synthetase - hyperuricemia gout excessive accumulation of uric acid in body fluids; arthritis pain in joints urate crystals in cartilage around joint; kidney stones Classification and causes of gout - primary inherited disorder - secondary induced by many disorders, leukemia - linked to defects in metabolism of purines; PRPP synthetase superactive variant associated with increased Vmax; another variant has an increased affinity (low Km) for ribose-5-P leading to overproduction of PRPP - 3rd defect associated with loss of feedback inhibition of this enzyme by purine nucleotides; when purine nucleotides reach excessive concentration no signal for shutting off their further production - defects of HGPRT inability to salvage purine bases from degradation overproduction of uric acid gout Gout treatment - allopurinol competitive inhibitor of xanthine oxidase; hypoxanthine and xanthine excreted during allopurinol therapy since xanthine oxidase uses both of these purines as substrates - allopurinol (like purine bases) can be converted to ribonucleotide form by HGPRT; treatment uses additional PRPP amidotransferase to reduce purine biosynthesis; analogue ribotide produced may inhibit PRPP amidotransferase - high [hypoxanthine] results from inhibition of xanthine oxidase causes HGPRT to reutilizes this base and further inhibit de no purine synthesis; purine synthesis lowered during allopurinol treatment - avoid animal products rich in nucleic acid (organ meats) Lesch-Nyhan syndrome - tremendous overproduction of uric acid; severe defect in HGPRT; males (X-linked) - aggressive behavior, mental retardation, self-mutilation - enzyme has essential role in non-hepatic tissue where de no synthesis of purines is slow non-hepatic tissues depend on circulating purine bases or nucleosides from liver - non-hepatic tissues thought to take up circulating purines and through HGPRT form nucleotides 2. PYRIMIDINE METABOLISM Pyrimidine biosynthesis and its regulation a. - glutamine (nitrogen donor) carbamoyl phosphate through carbamoyl phosphate synthetase II; urea cycle carbamoyl synthetase I uses ammonia - genetic defect of ornithine transcarbamoylase, in urea cycle, causes accumulation of carbamoyl phosphate that leaks from mitochondria to cytoplasm increase pyrimidine synthesis; increased pyrimidines excreted into urine (diagnose urea cycle defects) - pyrimidine biosynthesis continues with addition of aspartate via aspartate transcarbamoylase to provide remainder of ring elements carbamoyl aspartate - final product of pyrimidine synthesis is UMP can be phosphorylated to UTP cytidine triphosphate (CTP) - pyrimidines and ATP/GTP required for RNA synthesis b. - regulation of pyrimidine biosynthesis occurs at carbamoyl phosphate synthetase II feedback inhibition by uridine nucleotides (UDP, UTP) - PRPP in excess (PRPP, ATP) enzyme stimulated - to maintain balance of purine and pyrimidine nucleotides high amounts of purine nucleotides activate carbamoyl phosphate synthetase II 3. Metabolism of deoxyribionucleotides Formation of deoxynucleotides - 2-hydroxyl group of ribonucleotides reduced to form deoxyribonucleotide; catalyzed by ribonucleotide reductase; requires thioredoxin as a reducing source (it is oxidized); thioredoxin must be reduced back to its active form action of thioredoxin reductase (requires NADPH) - feedback regulation by deoxynucleotide triphosphates, dATP and dGTP Formation of thymidine - thymidine nucleotides required for DNA synthesis in place of UTP - TTP derived from TMP, which is formed from dUMP - dUMP TMP catalyzed by thymidylate synthase; dUMP acquires a carbon from N5,N10methyleneTHF converted to DHF 4. Metabolism of Folic Acid - folic acid/folate undergoes activation and conversion to various forms used in several biochemical reaction - folate serves as a donor of one carbon group - ingested folate converted to dihydrofolate reduced to tetrahydrofolate (THF) by dihydrofolate reductase - THF is backbone for production of other active forms of folate; must be in this form to carry carbon - N5,N10-methyleneTHF gains a carbon from side chain of serine yielding glycine product; required for TMP synthesis from dUMP - N5,N10-methenylTHF is a precursor for formation of N10-formyl THF required in purine biosynthesis (IMP formation); it is also a precursor of N5-methyl THF (in processing vitamin B12, cobalamin) provides methyl group for formation of methyl cobalamin; B12 deficiency folate trapped at N5-methyl THF because reaction from N5,N10-methyleneTHF is not reversible - folate supplementation prevents neural tube defects; prevent vascular disease 5. Chemotherapy and inhibition of DNA formation - cancer cells require DNA synthesis, chemotherapeutic agents inhibit DNA synthesis at level of synthesis of thymidine nucleotides because they are used selectively in DNA synthesis Flurodeoxyuridylate (F-dUMP) – is the product of metabolism of 5-fluorouracil (form of drug given) via orotate phosphoribosyl transferase; competitive inhibitor of thymidylate synthase Methotrexate – inhibits dihydrofolate reductase resulting in decreased synthesis of THF (DHF THF via DHF reductase normally); inhibitor ultimately decreases production of active forms of folate used in purine and pyrimidine synthesis (DNA synthesis disrupted) - analogues of folate given during chemotherapy to supply demand of slowly dividing normal cells