PathBinder User Manual
advertisement

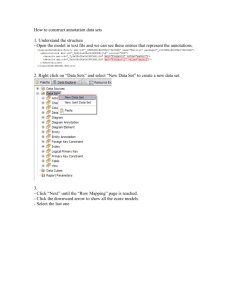
Documentation for PathBinder in MetNet Table of Contents (revise… 1 System Overview ........................................................................................................ 1 2 User Manual ................................................................................................................ 2 2.1 System Requirement ........................................................................................... 3 2.2 Installation Guide ...............................................Error! Bookmark not defined. 2.3 PathBinder Updater ............................................................................................. 7 2.3.1 2.3.2 2.3.3 2.3.4 2.4 2.4.1 3 Download MEDLINE files ............................................................................. 8 Update sentence repository ......................................................................... 9 Update species filters ...................................................................................... 9 Update dictionary ...................................................................................... 11 PathBinder Viewer .............................................................................................. 3 User Interface .............................................................................................. 3 Programmer Guide ...................................................................................................... 7 3.1 Database schema ............................................................................................... 12 3.2 Call PathBinder from Other Applications......................................................... 13 1 System Overview MEDLINE PathBinder Updater PathBinderDB in MetNet MetNetDB PathBinder Viewer FCModeler Figure 1. System overview of PathBinder in MetNet PathBinder is a gateway that provides enhanced access to MEDLINE information about metabolic interactions. MEDLINE baseline release files, as well as monthly update files, are searched against a precompiled dictionary, which consists of biochemical entities (metabolites, enzymes, co-factors, etc.), interaction-related verbs and sub-cellular locations. The sentences containing at least two search terms are stored in PathBinderDB, accessible through a GUI or programming API. Major enhancement includes: More relevant information retrieval. A user can query for sequences containing two biochemical entities, optionally in conjunction with interaction-related verbs or sub-cellular locations. Since the entities appear in the same sentences, they are more likely to have interactions with each other. On the other hand, if such queries were sent to PubMed directly, the entities might appear anywhere within an abstract, possibly far apart from each other with little chance of having interactions. Automatic synonym expansion. Any query term is automatically expanded to include its synonyms. Taxonomy filter. PathBinder provides a hierarchical taxonomy filter function, which enables a user to focus the queries on only the abstracts that mention the species of interest. 1 Combined with MetNetDB and FCModeler, PathBinder provides users with literature evidence for curating and simulating metabolic and regulatory pathways. PathBinder consists of three sub-systems, PathBinderUpdater, PathBinderViewer and PathBinderDatabase (a part of the MetNet database: http://www.public.iastate.edu/~mash/MetNet/MetNet_db.htm). PathBinderUpdater scans the entire MEDLINE release and monthly update off-line against a pre-compiled dictionary for sentences containing at least two search terms, and their synonyms. The search terms include biochemical entities (i.e. metabolites, proteins, and RNAs, and their synonyms, imported from the MetNetDB database), interactionrelated verbs (and their inflections), and selected sub-cellular locations (see table 1 for number of terms in each categories). PathbinderUpdater also collects species information from all abstracts by querying PubMed. PathBinderUpdater updates PathBinderDB every 2 or 3 months. The search and query results are stored in PathBinderDB. As of February 16, 2016, it contains over 17 million scanned sentences. See Programmer’s Guide for more details about its internal structure. PathBinderViewer is a stand-alone easy-to-use GUI for intuitive access of the information in PathBinderDB. The PathBinderViewer search tool also provides an API that can be evoked from other applications, such as FCModeler and MetNetDB. Thus, the user can be using software such as FCModeler or MetNetDB, click on any two terms on the screen, and evoke a PathBinderViewer window containing those two terms. Table 1. Statistics of PathBinderDB (February 16, 2016) Biochemical entities and synonyms Verbs and inflections Sub-cellular locations and synonyms Stored sentences 114,286 432 76 17,586,396 2 2 User Manual: PathBinderViewer--a biologists search tool This chapter describes how to install and use PathBinderViewer as a stand alone application. 2.1 System Requirement To use PathBinderViewer, a user needs a computer running Windows, Mac Os, Linux or Unix with Java J2SE 1.4 (or higher) and Java WebStart installed. A web browser and Internet connection are also required. 2.2 Start PathBinderViewer Type the following URL into a web browser’s address bar: http://metnetdb.gdcb.iastate.edu/PbFrame.jnlp If this is the first time running PathBinderViewer on the computer, Java WebStart will download PathBinderViewer program automatically from the server. After download is finished, WebStart will ask the user for permission to access local hard disk and Internet connection. Please click “Yes”. WebStart will also ask users whether or not to put a shortcut on the desktop. If a shortcut is created, PathBinderViewer can be started directly by double-clicking the shortcut, without the need to open a web browser and type in the URL. If this is not the first run, WebStart will check for updates to the program, and automatically download the updates if there is any. No user intervention is required. This makes sure that a user is always using the latest version of the program. 2.3 Users guide to PathBinder Viewer-a biologists search tool 2.3.1 User Interface The PathBinderViewer user interface consists of two separate windows: the control window (Fig. 2) and the sentence window (Fig. 3). The control window is for specifying the search terms (entities, verbs, and/or locations) and filters (taxonomy and/or location). The difference between a term and a filter is that the term must be present in a sentence, while a filter can appear anywhere in an abstract. For example, if a user specifies PKC and acetyl-coA carboxylase as two search terms, and Arabidopsis as a filter, PathBinderViewer will retrieve sentences containing both PKC and acetyl-coA carboxylase from the abstracts that contain Arabidopsis anywhere within the abstracts. The retrieved sentences are displayed in the sentence window with query terms highlighted. In addition, the synonyms of entity terms are listed. 3 Figure 2. PathBinderViewer: the control window The upper half of the control window is for setting up the taxonomy filter; and the lower half for specifying biochemical entities, verbs and/or sub-cellular locations. The entities and the verbs can only be used as search terms, while the locations may work in either term or filter mode. 2.3.2 Setup taxonomy filter There are three options for using the species filter: No filter (all abstracts are qualified). Use the simplified taxonomy tree in the upper-left window for commonly studied species of green plants. Hold down “Shift” or “Ctrl” key while clicking tree nodes to setup a filter including multiple species. Search the NCBI taxonomy database for plant species that are not present in the simplified tree. Type the keyword(s) in the “search term” field, and click the search button ( ) in the “Taxonomy search result” panel. Check the box(es) below the search field if you want an exact and/or case-sensitive search. By default, search result will overwrite the previous searches. However, if the “append” checkbox is checked, it will append the new search to the previous ones. The species in the search result list (upper-right window) can be removed with “delete” ( ) or “clear” ( ) commands, sorted ( ), and/or saved to a file 4 Figure 3. PathBinderViewer: sentence window ( ) for future use ( ). Highlight the items in the result list to setup the taxonomy filter (hold down “Shift” or “Ctrl” key while clicking an item for multiple selections). After highlighting the species names of interest in the simplified tree or in the search result, click the “Update filter” button. If sub-cellular locations are used in filter mode, also select the locations, and then click the “Update filter” button. The number of abstracts that pass the filter will be displayed as “PMID count.” 2.3.3 Specify search terms A user may ask PathBinder to show sentences that contain three types of search terms: One or more interaction-related verbs (and their reflections), optional; One or more sub-cellular locations (and their synonyms), optional; One or two entity names. If neither verbs nor locations are requested, two entities are required. Otherwise, at least one entity is required. Sub-cellular locations may also work in filter mode. Verb(s) and location(s) are organized into hierarchical groups in the lower-left and lowerright windows (Fig. 2). A single verb (location) or a group of verbs (locations) can be easily specified by clicking a tree node. Hold down “Shift” or “Ctrl” key while clicking 5 a node for multiple selections. Either tree may be quickly disabled by unchecking the box above the tree. There are three ways to populate the entity list (“Entity 1” and “Entity 2”): Searching in MetNet database’s “Entity” and “Entity_synonym” tables. . Type the keyword(s) in the “search term” field, and click the search button ( ) in the “Entity 1” or “Entity 2” panel. Check the box(es) below the search field for exact and/or case-sensitive search. The search result may overwrite or append to the previous searches (“append” checkbox). The list may be saved ( ) for future use. Loading entity names from a file ( ). The file may be previously saved search results, generated by other programs (MetNetDB, FcModeler, etc.), or manually created in any text editor. The file is in plain text format, 1 entity/line. Each line contains two fields separated by a vertical bar (“|”). The 1st field is EntityID in the “Entity” or “Entity_synonym” table, and the 2nd entity name. Inserted directly from other programs (MetNetDB, FcModeler, etc.). See “Programmer’s Guide” for more details. “Entity 2” list has an additional populating method by copying from “Entity 1” list ( ). Entities in both lists can be removed with “delete” ( ) or “clear” ( ) commands. The lists may be sorted ( ) alphabetically in ascending order. 2.3.4 Use sub-cellular locations as a filter By selecting the corresponding radio button, a user can use sub-cellular locations in filter mode. After selecting the location node(s), as well as species names if taxonomy filter is also used, click the “update filter” button. 2.3.5 Display sentences After setting up the species/location filter and specifying the search terms, click the “Show sentences” button to bring up the sentence window (Fig. 3). The requested sentences are displayed in the main window with search terms highlighted in different colors. All of the synonyms of entity 1 and/or entity 2 are shown in the drop-down lists above the main window. The PMID at the beginning of each sentence is a clickable link, which will open the corresponding PubMed abstract in a web browser. 6 3 System Administrators’ Guide to PathBinderUpdater This chapter describes how to install and use PathBinderUpdater as a stand alone application. PathBinderUpdater is used by the MetNetDB system administrator to keep PathbinderDB update-to-date with regard to MEDLINE. A frequency of updating every 2 or 3 months is recommended. PathBinderUpdater has four main functions: 1. Download MEDLINE XML release files and monthly updates; 2. Scan downloaded MEDLINE files against a dictionary to update the sentence repository; 3. Query PubMed to update species/location filter information; 4. Rebuild the dictionary if necessary (i.e. many changes have been made to the “Entity” and “Entity_synonym” tables in MetNet database). 3.1 System Requirement and Installation PathBinderUpdater has already installed on the server (metnetdb.gdcb.iastate.edu). No further installation is required. In case that the server is moved to a new computer, follow the steps below to setup PathBinderUpdater on the new server. The new server must have Java (J2SE 1.4 or higher) installed. There is no preference for operating systems. Copy the content of entire “c:\pb” fold (including sub-folds) to the new server. cleaned.tax Green plant taxonomy dictionary pb.dic Search term dictionary pb_metnet_src.jar Java source code pbframe.bat PathBinderViewer executable pbupdater.bat PathBinderUpdater executable sample_entity.list Sample entity list sample_species.list Sample species list singles_no_abb.txt Plural noun dictionary stop.list Stopwords for term dictionary lib\icons.zip Icons lib\liquidlnf.jar Liquid L&F library lib\pb_metnet.jar PathBinderUpdater library lib\pbframe.jar PathBinderView library Register the new server’s IP address at the MEDLINE FTP server, which can be requested via email by the licensee of MEDLINE. [Visit http://www.nlm.nih.gov/bsd/licensee.html for more details about licensing MEDLINE and registering IP address] 7 3.2 Download MEDLINE files remote folder file type filter remote folder content local folder local folder browser local folder content Figure 4. PathBinderUpdater user interface (MEDLINE FTP client) Users can browse files in four folders on the remote FTP server: Baseline gz (MEDLINE baseline release files compressed in gzip format) Baseline zip (MEDLINE baseline release files compressed in zip format) Monthly update (MEDLINE monthly update files in gzip and zip format) Sample (sample MEDLINE files) A user may select a file type filter to show only a certain type of files (zip only, gz only, or all type) in the remote folder content window. There are several methods to select the files for download: Click the file directly to select a single file; Press and hold down “Shift” or “Ctrl” key while clicking a file to select multiple files; Use the “Select all” button to select all files shown in the remote folder (files masked by the file type filter are not selected); Use the “Select undownloaded” button to select all files shown in the remote folder that are not in the local folder. Once one or more remote files are selected, the “download” button will be enabled. Clicking the button will start downloading process. Downloading and verifying md5 checksum files is optional. It is designed for unreliable network connections. If a file did not pass md5 check, it should be re-downloaded. However, during the process of downloading 619 baseline and monthly update files, there was not any mismatch. 8 By default, the downloaded MEDLINE files will be stored in the current working folder on the local computer. The local folder can be changed by using “local folder browser” or typing directly in the text field. 3.3 Update sentence repository entity dictionary plural noun dictionary local MEDLINE folder Figure 5. PathBinderUpdater user interface: Sentence repository The controls on the “Sentence repository” tab are disabled if the computer has not been connected to MetNetDB. To connect to MetNetDB, type in the MetNet server name (metnetdb.gdcb.iastate.edu), database name (dj_metnet), username and password, and click “Connect MetNet” button. To scan MEDLINE sentences, PathBinderUpdater needs a term dictionary, a plural noun dictionary, and the folder name where MEDLINE files are stored. The default term dictionary is pb.dic, and the default plural dictionary is singles_no_abb.txt. To use other dictionaries other than the default, type the file path directly in the corresponding text field, or select one using the “…” button. The entity dictionary needs periodical updates when the “Entity” and the “Entity_synonym” tables in MetNet database have been changed significantly. There is no need to update the plural noun dictionary. The MEDLINE folder is where the downloaded MEDLINE release files are stored. Once a folder is specified (typed in or selected through “…”), PathBinderUpdater searches for files whose names end with .xml, .xml.gz, or .xml.zip in the folder, and list them in the “Unprocessed files” window. All of the files in this list will be scanned. If some of the files were scanned before, highlight them (hold down “Shift” or “Ctrl” button for multiple selection) and click “<” button to move them into the “Processed files” window. After scanning, the files will be automatically moved to the “Processed files” window. The “<<” button moves all files from the unprocessed 9 window to the processed window; and the “>” and “>>” buttons move the files in the opposite direction. The “Clear sentence repository” button deletes all of the scanned sentences in the database for rebuilding the repository. It should be used after the entity dictionary is updated. The “Delete duplicated sentences” button is for the cases when same abstracts are scanned multiple times. This may happen accidentally, or because there are some duplicated PMIDs in MEDLINE monthly update files. 3.4 Update filters The controls on the “Sentence repository” tab are disabled if the computer has not been connected to MetNetDB. To connect to MetNetDB, type in the MetNet server name (metnetdb.gdcb.iastate.edu), database name (dj_metnet), username and password, and click “Connect MetNet” button. To update filters, select (use “…” button) or type in the species dictionary filename and the location dictionary filename, and click “Update” button. The default dictionaries are cleaned.tax and location.dic. Optionally, a user can type in the date of last update, so that the program only updates the most recent MEDLINE since last update. However, there is not much significant saving on running time by specifying a date (only about 5 ~ 10%). If no date is given, the previous filters should be deleted with the “Clear” button before the update, or using the “Delete duplicate” button after the update. Using “Clear” is much faster than “Delete duplicate.” After the update, the user should rebuild the simplified taxonomy tree (“Build simplified tree” button). Figure 6. PathBinderUpdater user interface: Species 10 3.5 Update dictionary The controls on the “Dictionary” tab are disabled if the computer has not connected to MetNetDB. To connect to MetNetDB, type in host name, database name, username and password, and click “Connect MetNet” button. Since the entire sentence repository needs a complete rebuild after update of the dictionary, it should be conducted when necessary, i.e. when significant changes have been made to the “Entity”, the “Entity_synonym”, the “Location” and the “Location_synonym” tables in MetNetDB. To create new dictionaries, type their filenames in the corresponding fields. To overwrite existing dictionaries, type in the old filenames or select with “…” button. An optional stopword file can be specified to exclude those entity names that are also common English words. A sample stopword file is in the working directory (c:\pb), stop.list. Figure 7. PathBinderUpdater user interface: Sentence repository 11 4 Programmer’s Guide This chapter descries the internal structure of PathBinder database, and how to use PathBinder from other applications (e.g. MetNetDB, FCModeler). 4.1 Database schema PathBinder adds to the MetNet database 15 tables, which can be conceptually divided into 4 groups around the “pb_sentence” table (Fig. 8). The table stores the sentences that contain at least two search terms, and to which PMIDs they belong. The group of location (verb) tables registers which location (verb) appears in which sentence and the information about the locations (verbs). The group of entity tables registers which entity appears in which sentence, similar to the verb and location groups. However, unlike the other two groups, there is an additional intermediate layer of termID between sentenceID and entityID. EntityIDs are first mapped to termIDs (pb_term2entity), and then mapped to sentenctIDs indirectly through termIDs (pb_term_hits). Such design is due to high ambiguity in MetNet entity names Entities Entity_synonym Entity PK pb_term2entity pb_term EntityID PK TermID 1:n FK1 EntityID Synonym Name Type Organism Username Date Source 1:n FK2 FK1 1:n EntityID TermID term 1:n Locations pb_term_hits pb_location_hits Verbs pb_verb PK FK1 FK2 pb_verb_hits verb verb_group 1:n FK1 FK2 1:n verbID SentenceID pb_sentence PK pb_prefilter FK2 FK1 tax_simplified PMID taxId m:n FK2 FK1 taxId parentId common_name name left_id right_id orderId 1:n pb_loc2pmid 1:n location_synonym m:n m:n verbID variant name definition goid parentID 1:n PMID Sentence m:n pb_verb_variant locationID SentenceID SentenceID 1:n FK1 FK2 FK1 1:n verbID location 1:n PK locationID TermID SentenceID FK2 FK1 PMID locationID FK1 locationID synonym pb_tax2pmid 1:n FK1 FK2 PMID taxId green_plants_node 1:1 1:n PK green_plants_name taxId parentTaxId rank left_id right_id 1:n FK1 taxid name nameClass Taxonomy Figure 2. PathBinder database schema 12 (i.e. a name or synonym corresponds to multiple entityIDs), and its purpose is to save table space and improve query speed. If entityIDs were used directly to register hits in the sentences, any ambiguous entity names (synonyms) would have to keep multiple copies of records in the database, each copy for one entityID. If several ambiguous names were found in the same sentence, all possible combinations of the entityIDs would need a copy of the record. If the most three ambiguous names in the current version of MetNet happened to be in the same sentence, it would need more than 5,000,000 copies. Therefore, a unique termID is assigned to every unique name (synonym), and only 1 copy is needed to register a hit. The group of taxonomy tables stores which PMIDs are associated with which taxa nodes. Please note that some information in the “tax_simplified” and “tax_prefilter” are redundant. It is there for faster queries. 4.2 Call PathBinderViewer from Other Applications Although other applications may retrieve PathBinder sentences from the above-described tables directly, it is not recommended to do so unless it is absolutely necessary. PathBinderViewer provides API for other applications to use PathBinder easily. To integrate PathBinder into other applications, include “pb_metnet.jar” in the class path. A programmer may call PathBinderViewer from the control window or the sentence window. 4.2.1 Call PathBinderViewer from the control window The biggest advantage of evoke PathBinderViewer at the control window is its easy-touse and flexible interface for setting up species filter and specifying locations and/or verbs. To activate the control window, simply create a new instance of PbFrame, and call its show() method. import edu.iastate.jtm.util.MySqlConnector; import fcmodeler.pathbinder.gui.PbFrame; ... DbConnector db = new MySqlConnector("metnetdb.gdcb.iastate.edu", "dj_metnet", "dingjing", "dingjing"); PbFrame pbf = new PbFrame(db); pbf.pack(); pbf.show(); ... // // // // server database username password The species filter setup and location (verb) selection cannot be done from outside the control window. That’s the point of using the control window. Otherwise, the other option (evoke the sentence window directly) might be considered. On the other hand, 13 there are several ways to populate the two entity lists, including ones from outside the control window. Use PathBinderViewer’s built-in search capability to find entities. Read from entity files. The files can be generated by other applications. Insert entities into the lists directly from the other application using PbFrame’s addEntity() method, which makes PathBinder seemingly more integrated with the application. After setting up the species filter and specifying the entities, locations and/or verbs, the sentence window can be brought up by clicking the “Show sentence” button in the control window, or calling PbFrame’s showPathBinder() method. 4.2.2 Call PathBinderViewer from the sentence window PathBinderViewer can also be evoked from the sentence window, if the capability of species filter, verbs and locations is not necessary, or does not meet the requirements of the other applications. The following sample code shows how to evoke the sentence window. import edu.iastate.jtm.util.MySqlConnector; import fcmodeler.pathbinder.PathBinder; import fcmodeler.pathbinder.PbSentence; import fcmodeler.pathbinder.gui.PbViewer; ... /* Create an instance of PathBinder, and send query to database. */ DbConnector db = new MySqlConnector("metnetdb.gdcb.iastate.edu", // server "dj_metnet", // database "dingjing", // username "dingjing"); // password PathBinder pb = new PathBinder(db); // Sentence querier pb.sendQuery(eid1, // int, entityID of entity 1 eid2, // int, entityID of entity 2 location, // Set, locationIDs in String (could be null) verb); // Set, verbIDs in String (could be null) /* Retrieve sentences and entity synonyms */ PbSentence[] sens = pb.getSentence(true); // sorted on PMIDs Set term1 = pb.getEntity1TermID(); Set t1name = pb.getEntity1TermName(); Set term2 = pb.getEntity2TermID(); Set t2name = pb.getEntity2TermName(); /* Create an instance of PbViewer, and display the retrieved sentences */ PbViewer pbViewer = new PbViewer(); // The sentence window pbViewer.setSentences(sens, term1, term2, t1name, t2name, location, verb); pbViewer.pack(); pbViewer.show(); ... 14 Optionally, a species/location filter can be setup before sending query for sentences. import fcmodeler.pathbinder.TaxonomyFilter; ... /* Create an instance of PmidFilter. */ PmidFilter pf = new PmidFilter(db); pf.addTaxEntry(“33090”, 124376, 222641); //Add a entry for “green plants” pf.updateFilterTable(true, null); // Create the filter ... PathBinder pb = new PathBinder(db); // Sentence querier pb.useFilter(true); pb.sendQuery(... 15