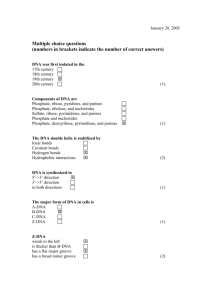
Supplementary files Figure S1: Semi
advertisement

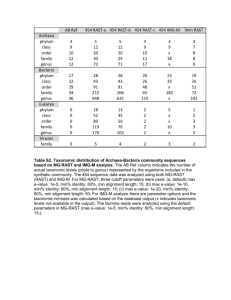
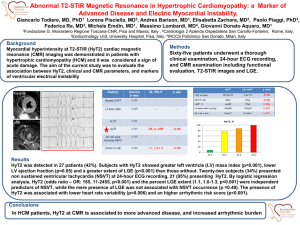
Supplementary files Figure S1: Semi-quantitative PCR of key melanin pathway genes in WT and Choc larvae. Semi-quantitative analysis of expression of the housekeeping gene FK506, seven genes of the melanin pathway, and candidate gene Ba_CSAD. Six individuals were examined for each stage and phenotype. The numbers of PCR cycles are shown on the right. LG assignment of the target genes in B. mori (cf. Silkworm Genome Database), together with primers and amplicon sizes, are given in Table S3. 1 Table S1: Segregation of Choc and mln individuals in experimental crosses. Family 1 2 3 4 5 6 7 8 Total Expected # (ratio) Number of F2 offspring with larval/adult phenotype Choc/mln Choc/WT WT/mln WT/WT 2 11 1 4 9 23 1 4 3 6 2 8 9 15 1 11 3 9 2 2 15 39 9 8 8 19 5 3 6 19 2 0 55 141 23 40 49 (3/16) 146 (9/16) 16 (1/16) Total offspring 18 37 19 36 16 71 35 27 259 49 (3/16) The numbers of F2 offspring are given for each of the four phenotypic classes (larval/adult phenotype). The difference between the observed total number of progeny were tested against the expected numbers if Choc and mln loci segregate independently with chi-square goodness-of-fit test (d.f. = 3). Table S2: Linkage between Choc and Ba_CSAD loci. Family Grandparents male female Choc 1A Choc (S/S) WT (M/L) Choc 1B Choc 2A Choc 2B WT (L/L) Choc (S/S) Parents male female Choc (S/M) Choc (S/L) Choc (S/M) Choc (S/L) Choc (S/L) Choc (S/L) Choc (S/L) Choc (S/L) Offspring Choc 29 S/S 24 M/L 30 S/M 17 S/L 21 S/S 19 M/L 21 S/M 32 S/L 36 S/S 29 L/L 15 S/L 23 S/S 13 L/L 40 S/L WT Total 100 93 80 76 The sizes of different alleles at the Ba_CSAD locus are indicated with S (1086-bp), M (1189-bp) and L (1305-bp). The genotypes of grandparents and parents are given in brackets. The numbers of F2 offspring of a particular genotype are given for WT and Choc phenotypes. 2 Table S3: B. anynana genes and primers used in semi-quantitative PCR. GenBank Amplicon Bmori Forward primer (5’- 3’) Reverse primer (5’- 3’) Accession size (bp) LG purple GE701593 CGGGAAACTTTCAGCTCTTG TGAAGAAACGTCATGGTGGT 455 5 pale GE740068 CAACCCTACCAGGACCAAGA GCACACAAGTCCCACGTCTA 339 1(Z) 316 Ddc GE713553 GACAAGTGGTGCCATCAGTG AACTCTTCGGGGAGTCCAAT 4* black JN003850 AATGGCGTATCCGTTAGCAA ACGTTGCACGCTATTCAGTG 260 11* tan GE686777 TCGCTTTTCCTCTTGCAAAT AATGGCTCATCAGCAAATCC 566 1(Z)* CSAD JN003848 ATTGACGCGTTCAAACTGTG AAATCACAACACAGGCAAGG 430 7* yellow GE715129 ACGGTGAAGCTGCAAGAAGT TACCCAACTCGTTTCCCTTG 274 13 ebony JN003849 CTTCCCACAAGGAAGGATCA GTGGAAAACCGCTCTCACAT 355 26 FK506 DY768120 CAAGACGGAGAAGTTCCACA AAACTAACCTGCAGCCCTGA 103 20* * Genes also mapped in B. anynana, all to orthologous LGs (Beldade et al., 2009). B. mori LG assignments follow Silkworm Genome Database (http://silkworm.genomics.org.cn/). Gene 3 Table S4: Predicted genes in the 1 Mbp interval of B. mori nscaf2986 around the orthologue of Ba_CSAD. Gene Best BLAST hit Position in nscaf2986 (size) BGIBMGA010134 BGIBMGA010248 BGIBMGA010133 BGIBMGA010249 BGIBMGA010132 BGIBMGA010250 BGIBMGA010251 BGIBMGA010131 BGIBMGA010252 BGIBMGA010130 BGIBMGA010129 E-value:4e-23 (EAT42776.1) conserved hypothetical protein [Aedes aegypti] E-value:1e-127 (XP_001359473.1) GA14898-PA [D. pseudoobscura] E-value:1e-49 (XP_971299.1) PREDICTED: similar to CG7390-PA, isoform A [T. castaneum] E-value:0.0 (EAT44265.1) conserved hypothetical protein [Aedes aegypti] E-value:1e-136 (XP_395226.2) PREDICTED: similar to CG14670-PA, partial [Apis mellifera] no hit E-value:1e-111 (NP_001040513.1) microtubule-associated protein 1 light chain 3 [B. mori] E-value:1e-101 (NP_001037105.1) intersex [B. mori] no hit E-value:0.0 (NP_001036841.1) Annexin IX-A [B. mori] E-value:0.0 (NP_001036911.1) ribonuclease L inhibitor homolog [B. mori] E-value:5e-25 (XP_001121740.1) PREDICTED: similar to Copper homeostasis protein cutC homolog [Apis mellifera] E-value:6e-35 (XP_001121520.1) PREDICTED: hypothetical protein [Apis mellifera] no hit no hit E-value:2e-14 (BAB40329.1) trypsinogen [Engraulis japonicus] E-value:1e-31 (XP_308309.3) ENSANGP00000010996 [Anopheles gambiae str. PEST] E-value:3e-19 (XP_641222.1) hypothetical protein DDBDRAFT_0206355 [Dictyostelium discoideum AX4] E-value:0.0 (AAC62238.1) NO-insensitive guanylyl cyclase [Manduca sexta] E-value:0.0 (XP_394067.2) PREDICTED: similar to Sema-5c CG5661-PA [Apis mellifera] no hit E-value:0.0 (NP_001040415.1) hemocyte protease [B. mori] E-value:1e-143 (XP_967423.1) PREDICTED: similar to CG5618-PA, isoform A [T. castaneum] E-value:1e-166 (XP_966646.1) PREDICTED: similar to soluble adenylyl cyclase [T. castaneum] E-value:1e-114 (XP_974494.1) PREDICTED: similar to CG32149-PA, isoform A [T. castaneum] E-value:0.0 (XP_394925.3) PREDICTED: similar to DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 [Apis mellifera] E-value:1e-145 (ABF18294.1) succinyl-CoA synthetase alpha subunit [Aedes aegypti] E-value:1e-171 (NP_649498.1) CG1129-PA, isoform A [D. melanogaster] E-value:0.0 (EAT36110.1) DNA mismatch repair protein muts [Aedes aegypti] E-value:1e-105 (EAT37426.1) zinc finger protein, putative [Aedes aegypti] E-value:1e-49 (XP_624045.2) PREDICTED: similar to Dynein light chain 90F CG12363-PA [Apis mellifera] E-value:1e-116 (XP_968497.1) PREDICTED: similar to CG5067-PA [T. castaneum] E-value:2e-24 (EAT44539.1) conserved hypothetical protein [Aedes aegypti] E-value:4e-29 (XP_971527.1) PREDICTED: similar to CG2663-PB, isoform B [T. castaneum] E-value:2e-28 (XP_970057.1) PREDICTED: similar to CG30339-PA [T. castaneum] no hit E-value:1e-08 (XP_970129.1) PREDICTED: similar to CG2663-PB, isoform B [T. castaneum] no hit E-value:3e-48 (NP_477435.1) fat-spondin CG6953-PA, isoform A [D. melanogaster] 3.306-3.309 Mbp (2.476 kbp) 3.311-3.321 Mbp (10.71 kbp) 3.327-3.331 Mbp (3.807 kbp) 3.337-3.363 Mbp (25.22 kbp) 3.368-3.381 Mbp (13.16 kbp) 3.382-3.382 Mbp (174 bp) 3.391-3.396 Mbp (5.144 kbp) 3.401-3.409 Mbp (7.615 kbp) 3.412-3.415 Mbp (2.181 kbp) 3.422-3.432 Mbp (10.22 kbp) 3.437-3.447 Mbp (10.3 kbp) BGIBMGA010253 BGIBMGA010128 BGIBMGA010127 BGIBMGA010126 BGIBMGA010254 BGIBMGA010255 BGIBMGA010256 BGIBMGA010125 BGIBMGA010124 BGIBMGA010123 BGIBMGA010257 BGIBMGA010122 BGIBMGA010258 BGIBMGA010121 BGIBMGA010259 BGIBMGA010120 BGIBMGA010260 BGIBMGA010261 BGIBMGA010119 BGIBMGA010262 BGIBMGA010263 BGIBMGA010118 BGIBMGA010117 BGIBMGA010116 BGIBMGA010115 BGIBMGA010114 BGIBMGA010264 BGIBMGA010265 3.452-3.452 Mbp (507 bp) 3.463-3.475 Mbp (12.36 kbp) 3.483-3.483 Mbp (279 bp) 3.514-3.517 Mbp (2.728 kbp) 3.572-3.572 Mbp (615 bp) 3.582-3.592 Mbp (10.52 kbp) 3.641-3.656 Mbp (14.44 kbp) 3.682-3.691 Mbp (8.562 kbp) 3.767-3.782 Mbp (15.17 kbp) 3.793-3.794 Mbp (1.429 kbp) 3.873-3.882 Mbp (8.983 kbp) 3.882-3.891 Mbp (8.31 kbp) 3.892-3.911 Mbp (19.34 kbp) 3.920-3.952 Mbp (31.61 kbp) 3.953-3.955 Mbp (1.971 kbp) 3.959-3.965 Mbp (6.218 kbp) 3.970-3.974 Mbp (3.879 kbp) 3.979-3.997 Mbp (17.96 kbp) 4.007-4.027 Mbp (19.52 kbp) 4.029-4.031 Mbp (1.652 kbp) 4.044-4.081 Mbp (37.06 kbp) 4.086-4.088 Mbp (1.941 kbp) 4.105-4.111 Mbp (6.087 kbp) 4.116-4.131 Mbp (15.07 kbp) 4.138-4.14 Mbp (2.213 kbp) 4.143-4.151 Mbp (7.625 kbp) 4.218-4.218 Mbp (237 bp) 4.303-4.307 Mbp (4.242 kbp) Gene annotation cf. Silkworm Genome Database; in darker grey – B. mori ortholog of Ba_CSAD, in lighter grey – putative genes in the interval presumably within 0.29 cM (corresponding to an estimated maximum of 174 Kb in B.mori) to the left and right side of it. 4