emi12086-sup-0011-TableS2
advertisement
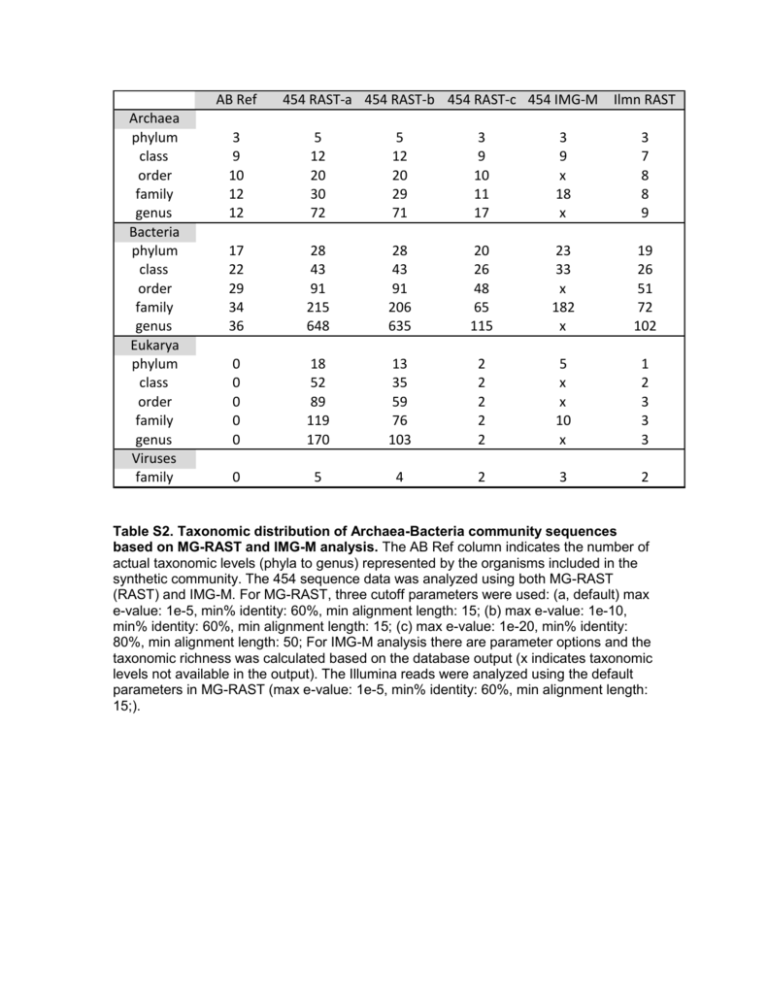
AB Ref Archaea phylum class order family genus Bacteria phylum class order family genus Eukarya phylum class order family genus Viruses family 454 RAST-a 454 RAST-b 454 RAST-c 454 IMG-M Ilmn RAST 3 9 10 12 12 5 12 20 30 72 5 12 20 29 71 3 9 10 11 17 3 9 x 18 x 3 7 8 8 9 17 22 29 34 36 28 43 91 215 648 28 43 91 206 635 20 26 48 65 115 23 33 x 182 x 19 26 51 72 102 0 0 0 0 0 18 52 89 119 170 13 35 59 76 103 2 2 2 2 2 5 x x 10 x 1 2 3 3 3 0 5 4 2 3 2 Table S2. Taxonomic distribution of Archaea-Bacteria community sequences based on MG-RAST and IMG-M analysis. The AB Ref column indicates the number of actual taxonomic levels (phyla to genus) represented by the organisms included in the synthetic community. The 454 sequence data was analyzed using both MG-RAST (RAST) and IMG-M. For MG-RAST, three cutoff parameters were used: (a, default) max e-value: 1e-5, min% identity: 60%, min alignment length: 15; (b) max e-value: 1e-10, min% identity: 60%, min alignment length: 15; (c) max e-value: 1e-20, min% identity: 80%, min alignment length: 50; For IMG-M analysis there are parameter options and the taxonomic richness was calculated based on the database output (x indicates taxonomic levels not available in the output). The Illumina reads were analyzed using the default parameters in MG-RAST (max e-value: 1e-5, min% identity: 60%, min alignment length: 15;).