Supporting Information - Springer Static Content Server
advertisement

Supporting Information
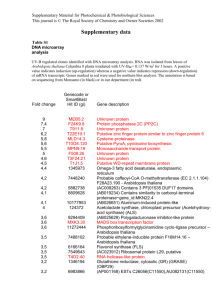
Table S1. Microarray expression analysis of ethylene-related genes (E-MEXP-254;
Siemens an others 2006)
TP1
TP2
description
AGI
0,67
0,69 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
1,30
0,51 2-oxoglutarate-dependent dioxygenase, putative identical to 2A6
AT1G03410
(GI:599622), a homolog of the tomato ethylene synthesis regulatory
protein E8; contains Pfam profile PF03171: oxidoreductase, 2OG-Fe(II)
oxygenase family
1,60
2,62 ethylene receptor-related similar to ethylene receptor CS-ETR2
[Cucumis sativus] GI:6136818; contains Pfam profiles PF01590:
GAF domain, PF00512: His Kinase A (phosphoacceptor) domain
AT1G04310
2,09
4,29 2-oxoglutarate-dependent dioxygenase, putative Similar to
Arabidopsis 2A6 (gb|X83096) and to tomato ethylene synthesis
regulatory protein E8 (SP|P10967)
AT1G04350
1,47
2,17 2-oxoglutarate-dependent dioxygenase, putative Strong similarity
to Arabidopsis 2A6 (gb|X83096), tomato ethylene synthesis
regulatory protein E8 (SP|P10967)
AT1G04380
1,51
2,56 1-aminocyclopropane-1-carboxylate oxidase / ACC oxidase /
ethylene-forming enzyme (ACO) (EAT1) Identical to 1aminocyclopropane-1-carboxylate oxidase (ACC oxidase)
gb|X66719 (EAT1).
AT1G05010
2,07
0,76 ethylene-responsive protein, putative similar to ethylene-inducible AT1G05710
ER33 protein [Lycopersicon esculentum] gi|5669656|gb|AAD46413;
identical to bHLH transcription factor (bHLH-alpha gene)
7,12
0,31 ethylene-responsive factor, putative similar to ethylene response
factor 1 GB:AAD03544 GI:4128208 from [Arabidopsis thaliana]
AT1G06160
1,17
0,42 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967); contains Pfam profile: PF00671 Iron/Ascorbate
oxidoreductase family
AT1G06620
0,51
0,25 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967); contains Pfam profile: PF00671 Iron/Ascorbate
oxidoreductase family
AT1G06640
0,67
2,50 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967); contains Pfam profile: PF00671 Iron/Ascorbate
AT1G06650
AT1G03400
TP1
TP2
description
AGI
oxidoreductase family
1,01
0,685 ethylene-responsive protein, putative similar to ER6 protein
AT1G09740
[Lycopersicon esculentum] GI:5669654; contains Pfam profile PF00582:
universal stress protein family
0,96
1,56 universal stress protein (USP) family protein contains Pfam PF00582:
universal stress protein family domain; similar to ethylene-responsive
ER6 protein (GI:5669654) [Lycopersicon esculentum]
1,08
AT1G26360
2,11 hydrolase, alpha/beta fold family protein similar to SP|Q40708
PIR7A protein {Oryza sativa}, ethylene-induced esterase [Citrus
sinensis] GI:14279437, polyneuridine aldehyde esterase [Rauvolfia
serpentina] GI:6651393; contains Pfam profile PF00561: alpha/beta
hydrolase fold
1,29
AT1G28160
2,09 ethylene-responsive element-binding family protein contains
similarity to ethylene-responsive element binding factor GI:8809573
from (Nicotiana sylvestris)
0,74
1,12 hydrolase, alpha/beta fold family protein similar to polyneuridine
AT1G33990
aldehyde esterase GI:6651393 from [Rauvolfia serpentina], SP|Q40708
PIR7A protein {Oryza sativa}, ethylene-induced esterase [Citrus
sinensis] GI:14279437; contains Pfam profile PF00561: hydrolase,
alpha/beta fold family
0,74
1,29 ethylene-responsive protein -related similarity to ER33 protein
[Lycopersicon esculentum] GI:5669656
AT1G49830
0,58
0,57 ethylene-responsive element-binding factor 3 (ERF3) identical to
SP|O80339 Ethylene responsive element binding factor 3 (AtERF3)
[Arabidopsis thaliana]
AT1G50640
1,18
1,17 ethylene-responsive element-binding factor 8 / ERF transcription factor 8 AT1G53170
(ERF8) identical to ERF transcription factor 8 GI:10567108 from
[Arabidopsis thaliana]
1,75
1,33 DEAD box RNA helicase, putative (RH20) similar to ethylene-responsive AT1G55150
RNA helicase GI:5669638 from [Lycopersicon esculentum]; contains
Pfam profiles PF00270: DEAD/DEAH box helicase, PF00271: Helicase
conserved C-terminal domain
AT1G11360
2,03
1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase,
putative nearly identical to ACC oxidase (ACC ox1) GI:587086 from
[Brassica oleracea]
AT1G62380
0,97
1,35 ethylene receptor 1 (ETR1) identical to GB:P49333 from [Arabidopsis
thaliana]
AT1G66340
0,31
0,42 calmodulin-binding protein similar to anther ethylene-upregulated
calmodulin-binding protein ER1 GI:11612392 from[Nicotiana
tabacum]
AT1G67310
2,20
AT1G69240
1,31 hydrolase, alpha/beta fold family protein low similarity to
SP|Q40708 PIR7A protein {Oryza sativa}, polyneuridine aldehyde
esterase GI:6651393 from [Rauvolfia serpentina], ethylene-induced
esterase [Citrus sinensis] GI:14279437; contains Pfam profile
TP1
TP2
description
AGI
PF00561: hydrolase, alpha/beta fold family
0,19
0,41 ethylene-responsive element-binding protein, putative contains
Pfam profile: PF00847 AP2 domain; similar to ethylene responsive
element binding protein (GI:18496063)[Fagus sylvatica]
0,57
0,49 ethylene-insensitive3-like3 (EIL3) identical to ethylene-insensitive3- AT1G73730
like3 (EIL3) GB:AF004215 [Arabidopsis thaliana]
1,46
AT1G79780
2,04 integral membrane protein, putative contains 1 transmembrane
domain; contains plant integral membrane protein domain,
TIGR01569; contains Pfam PF04535 : Domain of unknown function
(DUF588); similar to putative ethylene responsive element binding
protein (GI:22135858) [Arabidopsis thaliana]
0,76
2,05 ethylene-responsive element-binding family protein contains AP2
DNA-binding domain; similar to EREBP-3 (GI:1208496) [Nicotiana
tabacum]
AT1G80580
2,17
0,72 ethylene-responsive family protein similar to Ethylene-regulated
ER33 protein (GI:5669656) [Lycopersicon esculentum]; PMID:
12679534; putative bHLH133 transcription factor
AT2G20100
1,93
1,58 ethylene-responsive family protein similar to Ethylene-regulated ER33
protein (GI:5669656) [Lycopersicon esculentum]; PMID: 12679534;
putative bHLH133 transcription factor
AT2G20100
2,17
0,72 ethylene-responsive family protein similar to Ethylene-regulated
ER33 protein (GI:5669656) [Lycopersicon esculentum]; PMID:
12679534; putative bHLH133 transcription factor
AT2G20100
1,93
1,58 ethylene-responsive family protein similar to Ethylene-regulated ER33
protein (GI:5669656) [Lycopersicon esculentum]; PMID: 12679534;
putative bHLH133 transcription factor
AT2G20100
0,73
0,56 ethylene-responsive calmodulin-binding protein, putative (SR1) identical AT2G22300
to partial sequence of ethylene-induced calmodulin-binding protein
GI:11545505 from [Arabidopsis thaliana]; contains Pfam profiles
PF03859: CG-1 domain, PF00612: IQ calmodulin-binding motif, and
PF00023: Ankyrin repeat
2,33
0,51 hydrolase, alpha/beta fold family protein similar to ethyleneinduced esterase [Citrus sinensis] GI:14279437, polyneuridine
aldehyde esterase [Rauvolfia serpentina] GI:6651393; contains
Pfam profile PF00561: hydrolase, alpha/beta fold family
1,50
1,06 hydrolase, alpha/beta fold family protein similar to ethylene-induced
AT2G23560
esterase [Citrus sinensis] GI:14279437, polyneuridine aldehyde esterase
[Rauvolfia serpentina] GI:6651393; contains Pfam profile PF00561:
hydrolase, alpha/beta fold family
1,87
1,10 hydrolase, alpha/beta fold family protein similar to ethylene-induced
AT2G23570
esterase [Citrus sinensis] GI:14279437, polyneuridine aldehyde esterase
[Rauvolfia serpentina] GI:6651393
1,49
0,69 hydrolase, alpha/beta fold family protein similar to ethylene-induced
AT2G23580
esterase [Citrus sinensis] GI:14279437, polyneuridine aldehyde esterase
AT1G72360
AT2G23550
TP1
TP2
description
AGI
[Rauvolfia serpentina] GI:6651393; contains Pfam profile PF00561:
hydrolase, alpha/beta fold family
1,80
1,60 hydrolase, alpha/beta fold family protein similar to ethylene-induced
AT2G23590
esterase [Citrus sinensis] GI:14279437, polyneuridine aldehyde esterase
[Rauvolfia serpentina] GI:6651393; contains Pfam profile PF00561:
hydrolase, alpha/beta fold family
0,96
1,00 esterase, putative similar to ethylene-induced esterase [Citrus sinensis] AT2G23610
GI:14279437, polyneuridine aldehyde esterase [Rauvolfia serpentina]
GI:6651393; contains Pfam profile PF00561: hydrolase, alpha/beta fold
family
4,58
AT2G23620
2,21 esterase, putative similar to ethylene-induced esterase [Citrus
sinensis] GI:14279437, polyneuridine aldehyde esterase [Rauvolfia
serpentina] GI:6651393; contains Pfam profile PF00561: hydrolase,
alpha/beta fold family
1,46
3,02 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
1,28
1,29 ethylene-insensitive3-like1 (EIL1) identical to ethylene-insensitive3-like1 AT2G27050
GI:2224927 from [Arabidopsis thaliana]
1,34
0,95 expressed protein contains Pfam PF04535 : Domain of unknown
function (DUF588); similar to putative ethylene responsive element
binding protein (GI:22135858) [Arabidopsis thaliana]
AT2G28370
0,83
0,13 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
AT2G30830
1,84
0,59 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
AT2G30840
2,16
1,57 ethylene-responsive factor, putative similar to ethylene response
factor 1 GB:AAD03544 GI:4128208 from [Arabidopsis thaliana]
AT2G31230
0,69
0,59 ethylene-responsive protein, putative similar to ethylene-inducible ER33 AT2G31730
protein [Lycopersicon esculentum] gi|5669656|gb|AAD46413
1,70
1,55 integral membrane protein, putative contains 4 transmembrane domains; AT2G36330
contains Pfam PF04535 : Domain of unknown function (DUF588);
similar to putative ethylene responsive element binding protein
(GI:22135858) [Arabidopsis thaliana]
0,20
AT2G38210;A
2,60 [AT2G38210, ethylene-responsive protein, putative very strong
similarity to ethylene-inducible protein HEVER SP:Q39963 from
T2G38230;AT
[Hevea brasiliensis]];[AT2G38230, stress-responsive protein,
2G38240
putative similar to ethylene-inducible protein HEVER [Hevea
brasiliensis] SWISS-PROT:Q39963; contains Pfam domain,
PF01680: SOR/SNZ family];[AT2G38240, oxidoreductase, 2OG-Fe(II)
oxygenase family protein similar to flavonol synthase [Citrus
unshiu][gi:4126403], leucoanthocyanidin dioxygenase [Daucus
carota][gi:5924383]; contains PF03171 2OG-Fe(II) oxygenase
AT2G25450
TP1
TP2
description
AGI
superfamily domain]
0,65
0,58 integral membrane protein, putative contains 4 transmembrane domains; AT2G38480
contains plant integral membrane protein domain, TIGR01569 and
PF04535;: Domain of unknown function (DUF588); At3g16300,
At1g45222 both share this domain structure; distantly related to
GP|14030504 salicylic acid-induced fragment 1 protein {Gossypium
hirsutum}; similar to putative ethylene responsive element binding
protein (GI:22135858) [Arabidopsis thaliana]
1,20
AT2G38900
2,03 serine protease inhibitor, potato inhibitor I-type family protein
similar to SP|P24076 Glu S.griseus protease inhibitor (BGIA)
{Momordica charantia}, SP|P20076 Ethylene-responsive proteinase
inhibitor I precursor {Lycopersicon esculentum}; contains Pfam
profile PF00280: Potato inhibitor I family
0,99
1,65 integral membrane protein, putative contains 3 transmembrane domains; AT2G39530
contains Pfam PF04535 : Domain of unknown function (DUF588);
similar to putative ethylene responsive element binding protein
(GI:22135858) [Arabidopsis thaliana]
0,60
0,86 ethylene response sensor / ethylene-responsive sensor (ERS) identical
to ethylene response sensor (ERS) [Arabidopsis thaliana] GI:1046225
AT2G40940
1,22
1,17 ethylene-responsive transcriptional coactivator, putative similar to
ethylene-responsive transcriptional coactivator [Lycopersicon
esculentum] gi|5669634|gb|AAD46402
AT2G42680
0,87
0,85 late embryogenesis abundant family protein / LEA family protein similar
to ethylene-responsive late embryogenesis-like protein [Lycopersicon
esculentum] GI:1684830; contains Pfam profile PF03168: Late
embryogenesis abundant protein
AT2G44060
3,50
0,23 ethylene-responsive element-binding protein, putative
AT2G44840
1,21
1,10 NADH:ubiquinone oxidoreductase family protein contains Pfam
PF05071: NADH:ubiquinone oxidoreductase 17.2 kD subunit; similar to
ethylene-regulated ER6 protein (GI:5669654) [Lycopersicon
esculentum]; identical to Probable NADH-ubiquinone oxidoreductase
subunit B17.2 (EC 1.6.5.3) (EC 1.6.99.3) (Complex I-B17.2) (CI-B17.2)
(Swiss-Prot:Q9M9M9) [Arabidopsis thaliana]
AT3G03100
0,67
0,49 ethylene receptor, putative (EIN4) similar to ethylene receptor
GB:AAC31123 [Malus domestica], identical to putative ethylene
receptor GB:AAD02485 [Arabidopsis thaliana]; Pfam HMM hit:
response regulator receiver domain, signal C terminal domain
AT3G04580
0,79
1,28 hydrolase, alpha/beta fold family protein similar to ethylene-induced
esterase [Citrus sinensis] GI:14279437, SP|Q43360 PIR7B protein
{Oryza sativa}; contains Pfam profile PF00561: hydrolase, alpha/beta
fold family
AT3G10870
0,55
0,36 ethylene-responsive element-binding factor 4 (ERF4) identical to
ethylene responsive element binding factor 4 SP:O80340 from
[Arabidopsis thaliana]
AT3G15210
2,60
AT3G16050
0,75 stress-responsive protein, putative similar to ethylene-inducible
protein HEVER [Hevea brasiliensis] SWISS-PROT:Q39963; contains
TP1
TP2
description
AGI
Pfam domain, PF01680: SOR/SNZ family
0,70
1,07 calmodulin-binding protein similar to anther ethylene-upregulated protein AT3G16940
ER1 GI:11612392 from [Nicotiana tabacum]; contains Pfam profile:
PF00612 IQ calmodulin-binding motif (3 copies)
0,97
0,74 ethylene-responsive protein -related contains similarity to ethyleneinducible ER33 protein [Lycopersicon esculentum]
gi|5669656|gb|AAD46413
0,67
AT3G20310
0,46 ethylene-responsive element-binding family protein similar to
SP|O80339 Ethylene responsive element binding factor 3 (AtERF3)
{Arabidopsis thaliana}; contains Pfam profile PF00847: AP2 domain
0,69
0,90 ethylene-insensitive 3 (EIN3) identical to ethylene-insensitive3
GI:2224933 from [Arabidopsis thaliana]
0,82
1,94 ethylene receptor, putative (ETR2) similar to putative ethylene receptor; AT3G23150
ETR2 [Arabidopsis thaliana] gi|3687654|gb|AAC62208.
1,92
0,30 ethylene-responsive element-binding protein, putative similar to
SP:O80337,ERFI_ARATH Ethylene responsive element binding
factor 1 (AtERF1). {Arabidopsis thaliana}; similar to SP:O04681,
PTI5_LYCES Pathogenesis-related genes transcriptional activator
PTI5. [Tomato] {Lycopersicon esculentum} >GP|2213783|U89256;
similar to EREBP-2 GB:BAA07324 from [Nicotiana tabacum]
AT3G23220
0,98
0,84 ethylene-responsive factor, putative similar to EREBP-4 GB:BAA07323
from [Nicotiana tabacum]
AT3G23230
0,67
0,49 ethylene-responsive factor 1 / ethylene response factor 1 (ERF1)
identical to ethylene response factor 1 GB:AAD03544 from
[Arabidopsis thaliana]
AT3G23240
0,15
0,71 ethylene-responsive transcriptional coactivator, putative similar to AT3G24500
ethylene-responsive transcriptional coactivator [Lycopersicon
esculentum] gi|5669634|gb|AAD46402
5,47
1,09 hydrolase, alpha/beta fold family protein similar to ethyleneinduced esterase [Citrus sinensis] GI:14279437, polyneuridine
aldehyde esterase [Rauvolfia serpentina] GI:6651393; contains
Pfam profile PF00561: hydrolase, alpha/beta fold family
AT3G50440
1,34
2,00 integral membrane protein, putative contains 4 transmembrane
domains; contains Pfam PF04535 : Domain of unknown function
(DUF588); similar to putative ethylene responsive element binding
protein (GI:22135858) [Arabidopsis thaliana]
AT3G50810
0,96
1,88 ethylene-responsive transcriptional coactivator, putative similar to
ethylene-responsive transcriptional coactivator [Lycopersicon
esculentum] gi|5669634|gb|AAD46402
AT3G58680
3,07
AT3G61400
1,09 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967); 1-aminocyclopropane-1-carboxylate oxidase homolog
- Arabidopsis thaliana, PIR:S59548
AT3G19500
AT3G20770
TP1
0,91
3,51
TP2
description
AGI
1,31 late embryogenesis abundant 3 family protein / LEA3 family protein
AT4G02380
similar to several small proteins (~100 aa) that are induced by heat,
auxin, ethylene and wounding such as Phaseolus aureus indole-3-acetic
acid induced protein ARG (SW:32292); contains Pfam profile PF03242:
Late embryogenesis abundant protein
10,28 hydrolase, alpha/beta fold family protein similar to ethyleneinduced esterase [Citrus sinensis] GI:14279437, polyneuridine
aldehyde esterase [Rauvolfia serpentina] GI:6651393, SP|Q40708
PIR7A protein {Oryza sativa}; contains Pfam profile PF00561:
hydrolase, alpha/beta fold family
AT4G09900
2,11
0,90 translation elongation factor Ts (EF-Ts), putative similar to
ethylene-responsive elongation factor EF-Ts precursor
[Lycopersicon esculentum] GI:5669636; contains Pfam profile
PF00889: Elongation factor TS
AT4G11120
0,58
1,16 calmodulin-binding protein similar to anther ethylene-upregulated
calmodulin-binding protein ER1 GI:11612392 from [Nicotiana tabacum]
AT4G16150
1,07
3,34 esterase/lipase/thioesterase family protein similar to ethyleneinduced esterase [Citrus sinensis] GI:14279437, polyneuridine
aldehyde esterase [Rauvolfia serpentina] GI:6651393, SP|Q40708
PIR7A protein {Oryza sativa}; contains Interpro entry IPR000379
AT4G16690
0,91
AT4G17490
0,38 ethylene-responsive element-binding protein, putative similar to
SP|O80341 Ethylene responsive element binding factor 5 (AtERF5)
{Arabidopsis thaliana}
1,24
0,58 ethylene-responsive element-binding protein 1 (ERF1) / EREBP-2
AT4G17500
protein identical to SP|O80337 Ethylene responsive element binding
factor 1 (EREBP-2 protein) [Arabidopsis thaliana]; a false single bp exon
was added to circumvent a single basepair insertion in the genomic
sequence, supported by cDNA/genome alignment.
0,67
0,97 ethylene-responsive factor, putative similar to ethylene response factor 1 AT4G18450
GB:AAD03544 GI:4128208 from [Arabidopsis thaliana]; EREBP-1
(Ethylene-inducible DNA binding protein that interact with an ethyleneresponsive element) - Nicotiana tabacum, PATCHX:D1007899
1,21
0,95 ethylene-responsive nuclear protein / ethylene-regulated nuclear protein AT4G20880
(ERT2) identical to ethylene-regulated nuclear protein [Arabidopsis
thaliana] gi|2765442|emb|CAA75349
1,36
2,00 ethylene-responsive protein-related contains similarity to ethylene- AT4G21340
inducible ER33 protein [Lycopersicon esculentum]
gi|5669656|gb|AAD46413
2,19
0,92 ethylene-responsive family protein contains similarity to ethyleneinducible ER33 protein [Lycopersicon esculentum]
gi|5669656|gb|AAD46413
1,11
0,62 AP2 domain-containing transcription factor, putative ethylene-responsive AT4G34410
element binding protein homolog, Stylosanthes hamata, U91857
0,22
AT4G37150
1,14 esterase, putative similar to ethylene-induced esterase [Citrus
sinensis] GI:14279437, polyneuridine aldehyde esterase [Rauvolfia
serpentina] GI:6651393; contains Pfam profile PF00561: hydrolase,
AT4G29100
TP1
TP2
description
AGI
alpha/beta fold family
1,04
0,81 stress-responsive protein, putative similar to ethylene-inducible protein
HEVER [Hevea brasiliensis] SWISS-PROT:Q39963
AT5G01410
3,23
1,15 ethylene-insensitive 2 (EIN2) identical to EIN2 [Arabidopsis
thaliana] gi|5231113|gb|AAD41076; member of the natural
resistance-associated macrophage protein (NRAMP) metal
transporter family, PMID:11500563; metal transport capacity has
not been shown, PMID:11500563, PMID:1038174
AT5G03280
0,29
AT5G07580
2,70 ethylene-responsive element-binding family protein contains
similarity to ethylene responsive element binding factor 5 (AtERF5)
(Swiss-Prot:O80341) [Arabidopsis thaliana]; contains Pfam
PF00847: AP2 domain
0,72
1,16 calmodulin-binding protein similar to anther ethylene-upregulated
calmodulin-binding protein ER1 GI:11612392 from [Nicotiana tabacum]
AT5G09410
1,27
0,97 ethylene insensitive 3 family protein contains Pfam profile: PF04873
ethylene insensitive 3
AT5G10120
0,78
AT5G10300
0,43 hydrolase, alpha/beta fold family protein similar to ethyleneinduced esterase [Citrus sinensis] GI:14279437, alphahydroxynitrile lyase [Manihot esculenta] GI:2780225; contains Pfam
profile PF00561: hydrolase, alpha/beta fold family
0,89
2,24 ethylene-insensitive3-like2 (EIL2) identical to ethylene-insensitive3- AT5G21120
like2 (EIL2) GI:2224929 from [Arabidopsis thaliana]
2,10
4,00 ethylene-responsive element-binding protein, putative ethylene
responsive element binding protein homolog, Stylosanthes
hamata, EMBL:U91857
AT5G25190
2,85
1,37 integral membrane protein, putative MtN24 gene, Medicago
truncatula, EMBL:MTY15290; contains Pfam PF04535 : Domain of
unknown function (DUF588); contains 4 transmembrane domains;
similar to putative ethylene responsive element binding protein
(GI:22135858) [Arabidopsis thaliana]
AT5G40300
4,71
14,09 ethylene-responsive factor, putative contains AP2 DNA-binding
domain
AT5G43410
0,80
2,63 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
AT5G43440
2,88
7,08 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967)
AT5G43450
1,14
0,10 ethylene-responsive nuclear protein -related contains weak
similarity to ethylene-regulated nuclear protein ERT2 [Arabidopsis
thaliana] gi|2765442|emb|CAA75349
AT5G44350
2,01
AT5G47220
2,55 ethylene-responsive element-binding factor 2 (ERF2) identical to
SP|O80338 Ethylene responsive element binding factor 2 (AtERF2)
TP1
TP2
description
AGI
[Arabidopsis thaliana]
0,64
1,62 ethylene-responsive element-binding factor 5 (ERF5) identical to
SP|O80341 Ethylene responsive element binding factor 5 (AtERF5)
[Arabidopsis thaliana]
0,56
1,50 AP2 domain-containing transcription factor, putative contains similarity to AT5G51190
ethylene responsive element binding factor
0,82
AT5G58300;A
0,14 [AT5G58300, leucine-rich repeat transmembrane protein kinase,
putative];[AT5G58310, hydrolase, alpha/beta fold family protein low T5G58310
similarity to SP|Q40708 PIR7A protein {Oryza sativa}, polyneuridine
aldehyde esterase [Rauvolfia serpentina] GI:6651393, ethyleneinduced esterase [Citrus sinensis] GI:14279437; contains Pfam
profile PF00561: hydrolase, alpha/beta fold family]
1,22
2,95 hydrolase, alpha/beta fold family protein low similarity to
SP|Q40708 PIR7A protein {Oryza sativa}, polyneuridine aldehyde
esterase [Rauvolfia serpentina] GI:6651393, ethylene-induced
esterase [Citrus sinensis] GI:14279437; contains Pfam profile
PF00561: hydrolase, alpha/beta fold family
AT5G58310
0,45
0,33 2-oxoglutarate-dependent dioxygenase, putative similar to 2A6
(GI:599622) and tomato ethylene synthesis regulatory protein E8
(SP|P10967); 1-aminocyclopropane-1-carboxylate oxidase kidney
bean, PIR:T10818
AT5G59530
0,90
1,80 ethylene-responsive element-binding family protein contains similarity to AT5G61600
ethylene responsive element binding factor 5 (AtERF5) (SwissProt:O80341) [Arabidopsis thaliana]; contains Pfam PF00847: AP2
domain
0,78
0,81 ethylene-responsive DEAD box RNA helicase, putative (RH30) strong
AT5G63120
similarity to ethylene-responsive RNA helicase [Lycopersicon
esculentum] GI:5669638; contains Pfam profiles PF00270: DEAD/DEAH
box helicase, PF00271: Helicase conserved C-terminal domain
0,86
0,41 calmodulin-binding protein similar to anther ethylene-upregulated
calmodulin-binding protein ER1 GI:11612392 from [Nicotiana
tabacum]
AT5G64220
0,86
1,24 ethylene insensitive 3 family protein contains Pfam profile: PF04873
ethylene insensitive 3
AT5G65100
AT5G47230
Values of up- or down-regulated genes (>2 up-, <0.5 down-regulated) are highlighted
in red and blue, respectively, and the gene itself in bold. TP1 = 10 dai, TP2 = 23 dai.
Search term was “ethylene”. Genes are sorted according to their AGI number. Genes
further analyzed in this study are marked by * (transcript) and # (mutant).