ATF4 orchestrates a program of BH3
advertisement
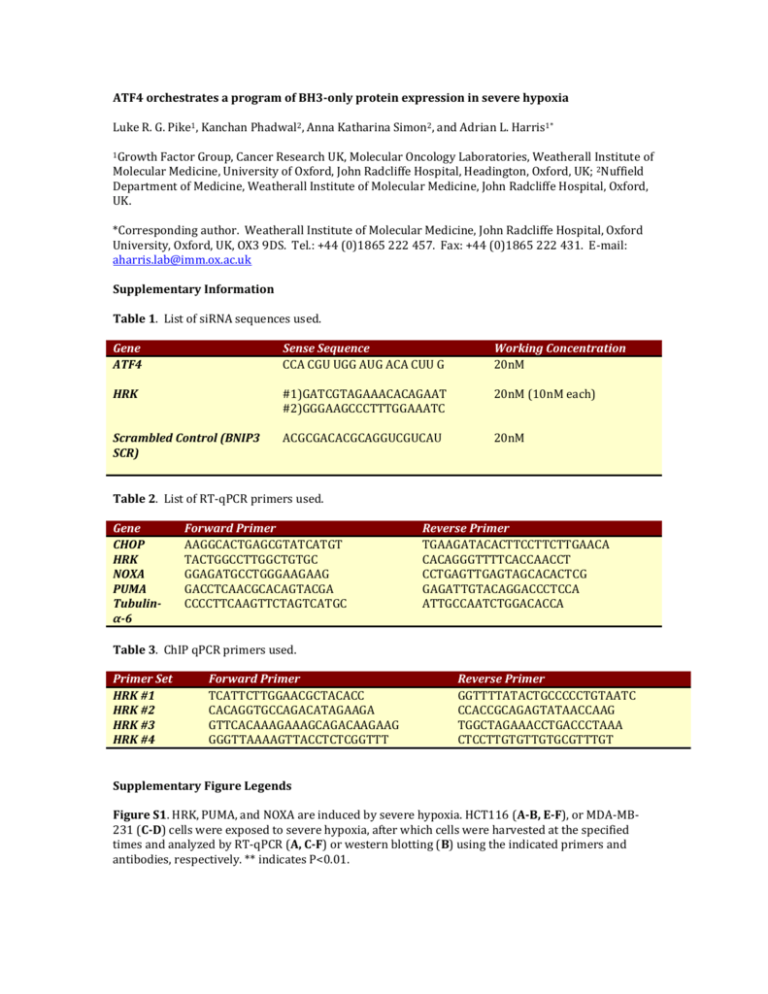
ATF4 orchestrates a program of BH3-only protein expression in severe hypoxia Luke R. G. Pike1, Kanchan Phadwal2, Anna Katharina Simon2, and Adrian L. Harris1* 1Growth Factor Group, Cancer Research UK, Molecular Oncology Laboratories, Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Headington, Oxford, UK; 2Nuffield Department of Medicine, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, UK. *Corresponding author. Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford University, Oxford, UK, OX3 9DS. Tel.: +44 (0)1865 222 457. Fax: +44 (0)1865 222 431. E-mail: aharris.lab@imm.ox.ac.uk Supplementary Information Table 1. List of siRNA sequences used. Gene ATF4 Sense Sequence CCA CGU UGG AUG ACA CUU G Working Concentration 20nM HRK #1)GATCGTAGAAACACAGAAT #2)GGGAAGCCCTTTGGAAATC 20nM (10nM each) Scrambled Control (BNIP3 SCR) ACGCGACACGCAGGUCGUCAU 20nM Table 2. List of RT-qPCR primers used. Gene CHOP HRK NOXA PUMA Tubulinα-6 Forward Primer AAGGCACTGAGCGTATCATGT TACTGGCCTTGGCTGTGC GGAGATGCCTGGGAAGAAG GACCTCAACGCACAGTACGA CCCCTTCAAGTTCTAGTCATGC Reverse Primer TGAAGATACACTTCCTTCTTGAACA CACAGGGTTTTCACCAACCT CCTGAGTTGAGTAGCACACTCG GAGATTGTACAGGACCCTCCA ATTGCCAATCTGGACACCA Table 3. ChIP qPCR primers used. Primer Set HRK #1 HRK #2 HRK #3 HRK #4 Forward Primer TCATTCTTGGAACGCTACACC CACAGGTGCCAGACATAGAAGA GTTCACAAAGAAAGCAGACAAGAAG GGGTTAAAAGTTACCTCTCGGTTT Reverse Primer GGTTTTATACTGCCCCCTGTAATC CCACCGCAGAGTATAACCAAG TGGCTAGAAACCTGACCCTAAA CTCCTTGTGTTGTGCGTTTGT Supplementary Figure Legends Figure S1. HRK, PUMA, and NOXA are induced by severe hypoxia. HCT116 (A-B, E-F), or MDA-MB231 (C-D) cells were exposed to severe hypoxia, after which cells were harvested at the specified times and analyzed by RT-qPCR (A, C-F) or western blotting (B) using the indicated primers and antibodies, respectively. ** indicates P<0.01. Figure S2. Severe hypoxia induces HRK expression in other cell lines. HEK293 (A), HeLA (B), MEF3T6 (C), and T47D cells (D) were exposed to severe hypoxia, after which cells were harvested and analyzed by RT-qPCR using primers specific for HRK. Figure S3. Chemical ER stressors induce HRK expression. (A) MCF7 cells were treated with 40M deferoxamine mesylate (DFO), 100M cobalt (II) chloride (CoCl2), 5M arsenite, 10M MG132, 5M MG115, 5g/mL tunicamycin, 300nM thapsigargin, 100nM bortezomib, or DMSO. After 24 hours, cells were harvested for RT-qPCR using primers specific for HRK. (B-E) Alternatively, four cell lines were exposed to 300nM thapsigargin and cells were harvested an analyzed by RT-qPCR using primers specific for HRK a the designated times. ** and * indicate P<0.01 or P<0.05 by one-wayANOVA relative to the DMSO treated control, respectively. Figure S4. HRK, PUMA, and NOXA are induced in an ATF4-dependent manner. MDA-MB-231 (A-D) or HCT116 cells (E-H) were transfected with siRNA against ATF4 (ATF4) or a scrambled control sequence (SCR) overnight and exposed to severe hypoxia (A) or normoxia (N) the following day. After 48 hours, cells were harvested for analysis by western blotting or RT-qPCR using the indicated antibodies and primers, respectively. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively. Figure S5. HRK expression in severe hypoxia is not HIF-dependent. MCF7 cells were transfected with siRNA molecules specific for HIF1 (1), HIF2 (2), or a scrambled control (SCR), and then exposed to normoxia, moderate hypoxia (H), or severe hypoxia (A). After 24 hours, cells were harvested and analyzed by RT-qPCR (A) or western blotting (B) using the specified primers and antibodies, respectively. For RT-qPCR, N=3 independent experiments were done. ** indicates P<0.01 by one-way-ANOVA for HIF2 A vs SCR A. For western blotting, representative images from at least N=2 independent experiments are shown. Figure S6. RNAi knockdown of HRK. MCF7 cells were transfected with a pool of sequence two siRNA duplexes specific for HRK at a final concentration of 20nM (10nM of each). 24 hours later, cells were exposed to severe hypoxia for a further 48 hours. Cells were then harvested and analyzed by RT-qPCR using primers specific for HRK. Figure S7. Knockdown of HRK reduces autophagy in HCT116 cells exposed to severe hypoxia (A) or thapsigargin (B). HCT116 cells were transfected with siRNA molecules specific for HRK or a scrambled control. After 24 hours, cells were exposed to normoxia (N), severe hypoxia (A) or treated with 300nM thapsigargin (TG) for an additional 24 hours and then harvested for western blotting using the indicated antibodies.











