Supplementary Discussion
advertisement

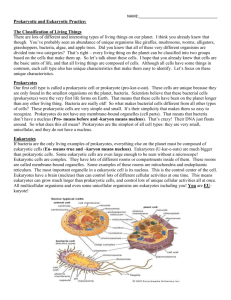
The Ring of Life Supplementary Discussion. Good methods are available for classifying tree-like phylogenies. But good methods for classifying the Ring of Life are currently lacking. For trees, node-based taxonomies effectively link classifications to phylogenies by creating a one-to-one correspondence between each node of a tree and all of the descendants from that node (including the node itself). Thus, if life evolves clonally, the nodes of a tree can define the classification. The phylogenetic groups produced by node-based procedures are naturally ranked so that the deepest nodes in the tree correspond to the most fundamental categories (domains, phyla, etc.)1. Since node based classifications can not be directly applied to graphs, we outline the procedures needed to apply node based classifications to the ring of life. The classification of rings is surprisingly straight forward, provided one breaks the ring into separate tree-like regions, and then applies the existing node-based procedures to each of the resulting regions, or realms. For example, consider the hypothetical tree shown in the figure above. Assume that the blue tree represents the prokaryotic realm and that the red tree represents the eukaryotic one. The prokaryotic realm comprises all prokaryotes and includes both of the fusion partners that created the eukaryote. These are represented as circles consisting of a blue and a red hemisphere. The eukaryotic realm comprises all of the eukaryotes and includes both of the fusion partners that founded the eukaryotes. By breaking the ring into trees, each realm may be classified separately, as if it were a tree. Yet, because the trees in both realms contain the fusion partners that close the ring, the separate classifications will retain all of the information that is necessary to reconstruct the ring. The method can also be applied to multiple rings. Phylogenetically-based ring classifications are simple to apply and fit common perceptions of natural groupings. For example, consider the eukaryotic and prokaryotic differences. Vastly different modes of cellular and genetic organization are found in eukaryotes and prokaryotes. Stanier et al. describe the differences as follows, “The numerous and fundamental differences between eukaryotic and prokaryotic organisms … probably represents the greatest single evolutionary discontinuity to be found in the present-day living world 2.” Not unexpectedly then, a node-based classification of the ring of life recognizes this fundamental difference between eukaryotes and prokaryotes and treats them as two different, but related, realms. At the same time, it also preserves the novel insights gained through the discovery of the Archaea. Domains Bacteria and Archaea could continue to be fundamental parts of the prokaryotes. We propose that the prefixes “Pro-” and “Eu-” currently used to distinguish prokaryotes from eukaryotes, should be combined with the suffix “Karya”, or “Carya” as used by Woese for eukaryotes in his pioneering contributions to the tree of life 3, to form the names of the realms, Prokarya and Eukarya , for the eukaryotic and prokaryotic realms, respectively. References 1. 2. 3. Sneath, P. H. A. in The Archaea and the deeply branching and phototrophic Bacteria. (eds. Boone, D. & Castenholz, R. W.) 83-88 (Springer, New York, Berlin, etc., 2001). Stanier, R., Adelberg, E. & Douderoff, M. The Microbial World. (Prentice-Hall, Englewood Cliffs, N.J., 1963). Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria and Eucarya. Proceedings of the National Academy of Sciences of the United 87, 4576-4579 (1990).