Appendix 3
advertisement

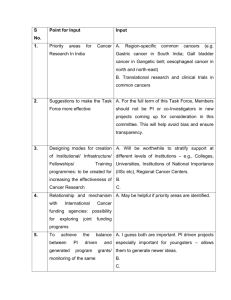
1. Top k-gene markers for distinguishing cancers of high survival rates Table 1 gives the top five one-gene special biomarkers across cancers of high survival rate. These genes can be used as biomarkers for the cancers of high survival rate, and simultaneously they are found to be effective discriminators for high survival rates and other survival rates cancers. S100B, KLHL21 and NAB1 are found to be used as the biomarkers for breast cancer. Similarly, MMD and NAB1 can be used as one-gene discriminator for prostate cancer. However, we didn’t find effective genes as biomarkers for thyroid cancer. The possible reason is that the samples number is small. The top five two-gene special biomarkers across cancers of high survival rate is showed in table 2. The combination of FBP1+S100B is found to be effective discriminator for three cancer types which have high survival rate. Similarly, the combinations of S100B+VSNL1 and ITGB1BP1+S100B can be used as two-gene discriminator for breast and thyroid cancers. Moreover, the combination of KLHL21+MMD is effective two-gene discriminator for breast and prostate cancers. Table 3 indicates the top five three-gene special biomarkers across cancers of high survival rate. The combinations of FBP1+S100B+VSNL1, MMD+NT5E+VSNL1 and CEBPB+FBP1+S100B are found to be effective discriminators for three cancer types which have high survival rate. Moreover, the combinations of KLHL21+MMD+VSNL1 and ARL4A+MMD+VSNL1 are good three-gene discriminator for breast cancer. The average classification accuracies of top 100 k-gene special markers are showed in figure 1. We also can see that the good 3-gene discriminators for cancer types of high survival rate have higher classification accuracies than the top 2-gene discriminators, and similarly the 2-gene discriminators are better than 1-gene. However, the best discriminative capacity of cancers which have high survival rate is not good enough. This may be because of the cancers of high survival rate have small number of specific genes in cancers formation and progression. As noted, S100B has been reported to play a key role in the regulation of a number of cellular processes such as cell cycle progression and also relate with survival rate of cancers [1], KLHL21 is suggested to have a possible role in cell division [2], MMD is involved in regulating ERK and Akt activation [3], LPIN1 is reported to have a possible role in p53 regulation [4], NAB1, which contains an active transcriptional repression domain, is reported as a co-repressor of EGR1 [5], and ITGB1BP1 is suggested to have a possible role in cell adhesion [6]. Moreover, several of the top genes as biomarkers have been reported to be cancer relevant. For example, FBP1 is reported to be down regulated in gastric carcinomas and used as a new biomarker for prognosis prediction of gastric cancer [7]. VSNL1 has been reporter to play a predictor role at the mRNA level of lymph node metastasis and poor prognosis in colorectal cancer [8]. CEBPB is suggested to play an important mediator of metalloproteinase gene activation during inflammatory responses in lung cancer cells [9]. Table 1: One-gene Marker for cancers of high survival rate Markers Breast Prosate Thyroid Accuracy1 Accuracy2 Mean S100B 76.84 60.56 68.33 70.85 66.78 68.81 KLHL21 74.27 67.40 49.58 67.91 69.40 68.65 MMD 67.16 71.24 56.67 66.36 69.94 68.15 LPIN1 61.75 69.72 67.08 64.90 69.17 67.03 NAB1 76.46 73.91 40.00 69.14 63.36 66.25 Table 2: Two-genes Marker for cancers of high survival rate Markers Breast Prosate Thyroid Accuracy1 Accuracy2 Mean FBP1+S100B 77.83 76.62 72.92 76.60 72.37 74.49 S100B+VSNL1 80.30 62.37 76.67 74.75 73.34 74.04 KLHL21+MMD 77.56 70.68 66.67 73.71 72.82 73.26 ITGB1BP1+S100B 77.77 66.92 75.83 74.46 71.84 73.15 MMD+VSNL1 77.59 67.50 62.08 72.02 73.67 72.84 Table 3: Three-genes Marker for cancers of high survival rate Markers Breast Prosate Thyroid Accuracy1 Accuracy2 Mean FBP1+S100B+VSNL1 81.94 75.38 77.92 79.42 76.15 77.79 KLHL21+MMD+VSNL1 82.92 70.40 66.67 76.55 78.26 77.40 MMD+NT5E+VSNL1 78.35 70.63 87.92 77.99 76.13 77.06 CEBPB+FBP1+S100B 76.03 76.31 77.92 76.45 76.85 76.65 ARL4A+MMD+VSNL1 80.25 68.94 76.25 76.44 76.70 76.57 1 1-gene 0.9 2-gene 3-gene 0.8 0.7 Accuracy 0.6 0.5 0.4 0.3 0.2 0.1 0 10 20 30 40 50 60 70 80 90 100 The number of genes Figure 1: Classification accuracies by the top 100 k-gene markers 2. Top k-gene markers for distinguishing cancers of medium survival rates Table 4 gives the top five one-gene special biomarkers across cancers of medium survival rate. These genes can be used as biomarkers for the cancers of medium survival rate, and simultaneously they are found to be effective discriminators for medium survival rates and other survival rates cancers. MTHFD1 and NEDD4L are found to be used as the biomarkers for colorectal cancer. Similarly, DOCK4 and C11orf9 can be used as one-gene discriminator for lung cancer. However, we didn’t find effective genes as biomarkers for gastric cancer. The possible reason is the various types of gastric cancer. The top five two-gene special biomarkers across cancers of medium survival rate is showed in table 5. The combinations of DOCK4+MTHFD1, EMP2+MTHFD1 and ABCG1+MTHFD1 are found to be good discriminator for lung and colorectal cancers. Similarly, the combinations of MTHFD1+NEDD4L and EDIL3+MTHFD1 can be used as two-gene discriminator for colorectal cancers. Table 6 indicates the top five three-gene special biomarkers across cancers of medium survival rate. The combinations of CAPN9+EMP2+MTHFD1 and CAPN9+DOCK4+MTHFD1 are found to be effective discriminators for three cancer types which have medium survival rate. Moreover, the combinations of EMP2+MTHFD1+NEDD4L, DOCK4+GSTP1+MTHFD1 and EMP2+MTHFD1+TRIM2 are good three-gene discriminator for lung and colorectal cancers. The average classification accuracies of top 100 k-gene special markers are showed in figure 2. We also can see that the good 3-gene discriminators for cancer types of medium survival rate have higher classification accuracies than the top 2-gene discriminators, and similarly the 2-gene discriminators are better than 1-gene. As noted, DOCK4 is involved in regulation of adherens junctions between cells and regulates intercellular junctions [10] TSPAN6 is reported to play a role in the regulation of cell development, activation, growth and motility [11]. ABCG1 has been suggested to play an important role in cellular lipid/sterol regulation [12]. TRIM2 has been suggested to play a role in mediating the p42/p44 MAPK-dependent ubiquitination [13] Moreover, several of the top genes as biomarkers have been reported to be cancer relevant. For example, MTHFD1 is reported to play a key role among folate metabolism and colon cancer initiation and progression [14]. NEDD4L has been reported to play a role in the progression of various cancers. The negative expression of the gene is strongly related to the invasion and metastasis of gastric cancer [15]. EMP2 is suggested to be related with endometrial and ovarian cancers [16, 17]. EDIL3 has reported to play a role in tumor angiogenesis and the interaction between hepatocellular carcinoma cells and endothelial cells [18]. The gene CAPN9 is reported to be suppressed or depleted with considerable frequency in gastric cancer [19]. GSTP1 has been suggested to relate with cell death and play a role in susceptibility to colon cancer [20]. Table 4: One-gene Marker for cancers of medium survival rate Markers Lung Colorectal Gastric Accuracy1 Accuracy2 Mean MTHFD1 73.55 80.46 56.71 71.30 76.75 74.03 NEDD4L 69.64 87.08 65.92 74.17 70.22 72.19 DOCK4 87.47 64.00 62.71 73.54 70.25 71.90 C11orf9 82.91 57.33 67.91 70.89 68.64 69.77 TSPAN6 73.24 68.75 66.89 70.15 68.76 69.46 Table 5: Two-genes Marker for cancers of medium survival rate Markers Lung Colorectal Gastric Accuracy1 Accuracy2 Mean DOCK4+MTHFD1 90.14 84.59 63.85 81.47 79.40 80.44 EMP2+MTHFD1 89.26 82.27 56.73 78.49 80.76 79.63 MTHFD1+NEDD4L 79.80 92.51 63.65 79.57 78.45 79.01 EDIL3+MTHFD1 77.61 89.60 64.03 77.82 80.09 78.95 ABCG1+MTHFD1 80.67 81.33 59.28 75.25 82.11 78.68 Table 6: Three-genes Marker for cancers of medium survival rate Markers Lung Colorectal Gastric Accuracy1 Accuracy2 Mean CAPN9+EMP2+MTHFD1 89.18 86.85 75.87 84.94 83.52 84.23 CAPN9+DOCK4+MTHFD1 89.72 90.81 72.73 85.59 82.74 84.17 EMP2+MTHFD1+NEDD4L 92.12 91.44 65.71 84.96 82.34 83.65 DOCK4+GSTP1+MTHFD1 91.73 85.07 70.07 83.92 82.53 83.23 EMP2+MTHFD1+TRIM2 93.97 82.23 70.58 84.11 82.17 83.14 1 0.9 0.8 0.7 Accuracy 0.6 1-gene 0.5 2-gene 3-gene 0.4 0.3 0.2 0.1 0 10 20 30 40 50 60 70 80 90 100 The number of genes Figure 2: Classification accuracies by the top 100 k-gene markers References [1] Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 2008;34:357-64. [2] Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. The Journal of cell biology 2009;187:791-800. [3] Liu Q, Zheng J, Yin DD, Xiang J, He F, Wang YC, et al. Monocyte to macrophage differentiation-associated (MMD) positively regulates ERK and Akt activation and TNF-alpha and NO production in macrophages. Molecular biology reports 2012;39:5643-50. [4] Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Molecular cell 2011;44:491-501. [5] Swirnoff AH, Apel ED, Svaren J, Sevetson BR, Zimonjic DB, Popescu NC, et al. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Molecular and cellular biology 1998;18:512-24. [6] Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Human molecular genetics 2002;11:389-96. [7] Liu X, Wang X, Zhang J, Lam EK, Shin VY, Cheng AS, et al. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene 2010;29:442-50. [8] Akagi T, Hijiya N, Inomata M, Shiraishi N, Moriyama M, Kitano S. Visinin-like protein-1 overexpression is an indicator of lymph node metastasis and poor prognosis in colorectal cancer patients. International journal of cancer Journal international du cancer 2012;131:1307-17. [9] Armstrong DA, Phelps LN, Vincenti MP. CCAAT enhancer binding protein-beta regulates matrix metalloproteinase-1 expression in interleukin-1beta-stimulated A549 lung carcinoma cells. Molecular cancer research : MCR 2009;7:1517-24. [10] Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell 2003;112:673-84. [11] Maeda K, Matsuhashi S, Hori K, Xin Z, Mukai T, Tabuchi K, et al. Cloning and characterization of a novel human gene, TM4SF6, encoding a protein belonging to the transmembrane 4 superfamily, and mapped to Xq22. Genomics 1998;52:240-2. [12] Cserepes J, Szentpetery Z, Seres L, Ozvegy-Laczka C, Langmann T, Schmitz G, et al. Functional expression and characterization of the human ABCG1 and ABCG4 proteins: indications for heterodimerization. Biochemical and biophysical research communications 2004;320:860-7. [13] Thompson S, Pearson AN, Ashley MD, Jessick V, Murphy BM, Gafken P, et al. Identification of a novel Bcl-2-interacting mediator of cell death (Bim) E3 ligase, tripartite motif-containing protein 2 (TRIM2), and its role in rapid ischemic tolerance-induced neuroprotection. The Journal of biological chemistry 2011;286:19331-9. [14] MacFarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ. Mthfd1 is a modifier of chemically induced intestinal carcinogenesis. Carcinogenesis 2011;32:427-33. [15] Gao C, Pang L, Ren C, Ma T. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Medical oncology 2012;29:1733-8. [16] Shimazaki K, Lepin EJ, Wei B, Nagy AK, Coulam CP, Mareninov S, et al. Diabodies targeting epithelial membrane protein 2 reduce tumorigenicity of human endometrial cancer cell lines. Clinical cancer research : an official journal of the American Association for Cancer Research 2008;14:7367-77. [17] Fu M, Maresh EL, Soslow RA, Alavi M, Mah V, Zhou Q, et al. Epithelial membrane protein-2 is a novel therapeutic target in ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16:3954-63. [18] Sun JC, Liang XT, Pan K, Wang H, Zhao JJ, Li JJ, et al. High expression level of EDIL3 in HCC predicts poor prognosis of HCC patients. World journal of gastroenterology : WJG 2010;16:4611-5. [19] Yoshikawa Y, Mukai H, Hino F, Asada K, Kato I. Isolation of two novel genes, down-regulated in gastric cancer. Japanese journal of cancer research : Gann 2000;91:459-63. [20] Muller J, Sidler D, Nachbur U, Wastling J, Brunner T, Hemphill A. Thiazolides inhibit growth and induce glutathione-S-transferase Pi (GSTP1)-dependent cell death in human colon cancer cells. International journal of cancer Journal international du cancer 2008;123:1797-806.