CcpNmrTutorial
advertisement
Course Organization
The course is organized in four parts:
1. Getting started; Display and navigation; Overview.
2. Molecules; Assigning peaks.
3. Efficient backbone and side chain assignment.
4. Different spectrum conditions; Relaxation analysis.
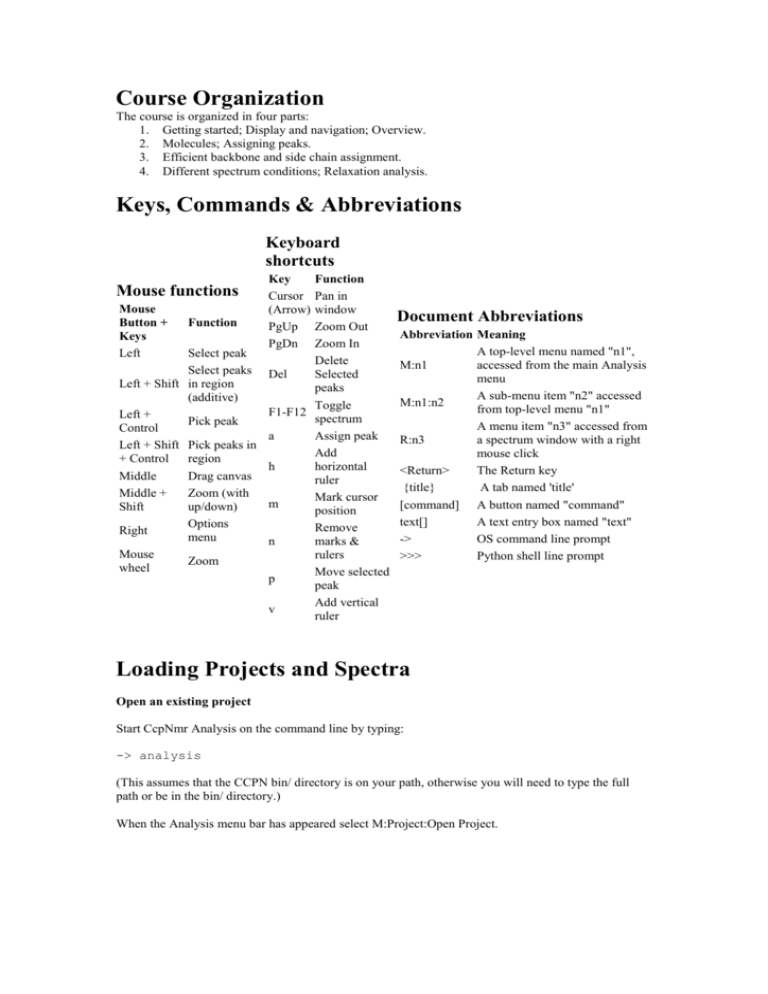
Keys, Commands & Abbreviations
Keyboard
shortcuts
Function
Pan in
Mouse
window
Button +
Function
Zoom Out
Keys
Zoom In
Left
Select peak
Delete
Select peaks Del
Selected
Left + Shift in region
peaks
(additive)
Toggle
F1-F12
Left +
spectrum
Pick peak
Control
a
Assign peak
Left + Shift Pick peaks in
Add
+ Control region
h
horizontal
Middle
Drag canvas
ruler
Middle +
Zoom (with
Mark cursor
m
Shift
up/down)
position
Options
Remove
Right
menu
n
marks &
Mouse
rulers
Zoom
wheel
Move selected
p
peak
Add vertical
v
ruler
Mouse functions
Key
Cursor
(Arrow)
PgUp
PgDn
Document Abbreviations
Abbreviation Meaning
A top-level menu named "n1",
M:n1
accessed from the main Analysis
menu
A sub-menu item "n2" accessed
M:n1:n2
from top-level menu "n1"
A menu item "n3" accessed from
R:n3
a spectrum window with a right
mouse click
<Return>
The Return key
{title}
A tab named 'title'
[command] A button named "command"
text[]
A text entry box named "text"
->
OS command line prompt
>>>
Python shell line prompt
Loading Projects and Spectra
Open an existing project
Start CcpNmr Analysis on the command line by typing:
-> analysis
(This assumes that the CCPN bin/ directory is on your path, otherwise you will need to type the full
path or be in the bin/ directory.)
When the Analysis menu bar has appeared select M:Project:Open Project.
Navigate to find and then select the CcpnCourse1a project (the directory initially will be in green and
then when you have selected it in deep purple).
Then click [Open].
You might get a warning that various files have moved location, which is fine; just click [OK].
Dependent on how the project files and directories have been unpacked, you might also get a dialogue
panel with a list of spectra paths (because these also have moved location). If any path is in red then
Analysis cannot find the corresponding spectrum data file, so either you need to tell Analysis where it
is (by double clicking the path cell and navigating to the correct location) or accept that that particular
spectrum will not have its contours displayed.
Now save the new project by selecting M:Project:Save. The first time you save a project that has
moved locations the SaveAs dialog is brought up. At this point you can choose to change the project
name and/or the location where the project directory will be saved.
It will highlight in red existing project directories, so that you do not accidentally overwrite one. Here
we are happy to save onto the existing project (we are just updating the paths). So just hit the "Save"
button, and also OK to the question about overwriting the existing project directory.
After this first save, M:Project:Save will just automatically save to the existing location.
M:Project:SaveAs will let you save your project with another name, or in another location, but will still
have the directory name the same as the project name It is a good idea to save now and again in case
something goes wrong (either with your work or with the code).
This project has two windows, "window1" and "window2". The first is a two-dimensional window
with an HSQC spectrum in it, and the second is a three-dimensional window with two spectra in it, an
HNCA and an HNcoCA.
Opening Spectra from File
To open a new spectrum go to M:Experiment:Open Spectra.
In the resultant Open Spectra popup navigate to the CcpnCourse1a/spectra/ directory and select
182.spc.par (the other files are spectra that are already open, or that will be needed later).You can open
up more than one spectrum at one time, but there we will just open one. If necessary, enlarge the
popup (click and drag the bottom edge down) so that you can see the entry in the bottom table. This
table is a list of how the selected spectra will be placed into the CCPN data structure. Each file will be
associated with an experiment and a spectrum that are in turn linked to a shift list. For the most part
each experiment carries only one spectrum, as will be the case in this tutorial. However, it is possible to
have one experiment with several spectra, e.g. when they are processed differently. All the spectra will
use the same shift list, as the experimental conditions are the same, so leave that field alone.
Enter a name for the selected experiment by double clicking on the experiment name cell, "Expt_4",
and entering the name and then pressing <Return> or clicking outside the box. Set the name of the
experiment to "N-NOESY" or similar. The spectrum name could also be altered, but this is not
necessary here and the spectrum name being inherited from the spectrum file ("182.spc") is
informative.
Note that the file format option at the top op the popup is set to "Azara", which is correct for the files
we will open. If the spectra were stored in a different format (Bruker, Felix, NMRPipe, NMRView,
UCSF, Varian or Factorised) this option would have to be altered.
Finally click [Open Spectrum].
Now file and referencing verification dialogues will appear. Again, enlarge the popup so you can see
the entire table. The {Verify Referencing} tab will be open - it shows the parameters you normally
need to look at.
The {Verify File Details} tab lets you edit where and how the file is stored, but the information here is
normally correct.
Click [Commit] on the button at the top of the popup (from either tab) to continue. Note that you can
change spectrum referencing and file setup at any time after loading via M:Experiment:Spectra.
Note that you may skip all the verification steps, if you know your parameters to be correct, by
selecting the "Skip verification dialogs" option before you click [Open Spectra]. You can change
almost all the spectrum referencing and file parameters at a later time should you need to make
adjustments. The only thing that you cannot change after a spectrum is loaded are its primary axes; in
terms of number and which isotopes they refer to.
Setting Experiment Types
After the verification there is one final popup where you specify the types of the experiment that was
run. This information is very useful later on; for example Analysis uses the knowledge of which
experimental dimensions correspond to single bond transfers to automatically remove impossible
assignment options.
Set the synonym for the new experiment to "15N HSQC-NOESY". Note that for this experiment there
are two different full types with the same synonym (double click the Full Type column). So the "15N
HSQC-NOESY" could be either H_H[N].NOESY or H[N]_H.NOESY, with the difference being that
the NOESY transfer can come either before or after the HSQC step. For our experiment the selection
H_H[N].NOESY is correct.
When the experiment type is set click [Done] at the top right or [Close - All Done] at the bottom. You
may have to expand the popup to see the [Done] button. After a few moments you will hopefully see
the newly loaded spectrum appear. You can now also close the Open Spectra popup.
Spectrum Window Navigation
Window Components and Views
Once the new spectrum has loaded, a new window, "window3" will automatically be created. This is a
three-dimensional window with "1H" along the x and y axes and "15N" along the z axis. A new
window is created like this only if no existing window matches in isotopes.
Note that if you minimise or close a window you can get it back again by going to M:Windows:HN:
window3 (or equivalent).
You can move around the spectra within the windows using several different inputs. To zoom in and
out use the <PgUp> and <PgDn> keys, the middle mouse wheel (if you have one), or hold <Shift>,
click the middle mouse button, and move the mouse up and down. If you zoom out from your spectra
so that you can see their edge you will notice a dotted line which denotes their border.
You cannot zoom out further than the maximum size allowed in each dimension. To change this size
open up M:Window:Axes and select the {Axis Types} tab (if it is not already selected). Each Axis
Type has a Region, which can be edited by double clicking on the relevant cell.
To pan around the spectra you can click and drag the scrollbars at the edges of the windows, use the
arrow keys, or click and drag on the spectra with the middle mouse button.
Looking in "window2" move the extra scrollbar at the very bottom of the window. - Left click and
drag. This scrollbar is present on the 3D window to change the depth or plane of the spectra being
viewed. On a 4D window there would be yet another scrollbar. To change the thickness of the
displayed planes click and drag the side of the depth slider with the middle mouse button.
Staying in window2, click on "Spectra" at the top of the window. Here you will see two coloured
buttons, one for each of the 3D spectra. If you click the buttons you can independently toggle the
contour displays for the two spectra on and off.
Setting Contours
To change contour settings click on the "Contours" button at the top of a window, try doing this in
window3 for the newly loaded spectrum. This lets you do simple adjustments. The green arrows will
raise or lower the contouring floor, while the +1/-1 will change the number of contours. The
"Pos/Neg" button allows you to swap between showing just positive or just negative or both contour
levels. Note that using these contour options only changes the spectra that are visible at the time within
the window. If you click on the "More..." button you will bring up a more detailed contour level setting
dialogue box.
Picking peaks
Peak Picking and Manipulation
Focusing on the HSQC spectrum in "window1", the next task is to define some contour extrema as
peaks. There are two common ways to pick peaks. One is to search for all extrema in a boxed region.
Try this in the HSQC window by holding down <Shift> and <Ctrl> whilst clicking with the left mouse
button and dragging the (dappled) box to define a rectangular pick region.
Note that when the peaks are picked the contours will carry diagonal crosses to denote the peak
position. Each peak will also have some annotation to the top right of the cross, which we discuss
below. Clicking on the screen
Locate the contours near the point at 8.7 ppm on the 1H axis and 119.0 on the 15N axis. These
contours represent two overlapping peaks where the extrema search will only pick one of the two
underlying signals. To define the second peak position, hold down <Ctrl> and click on the location to
pick the new peak.
Now we will select some of the peaks, e.g. artefacts or noise, for deletion. To delete peaks click with
the left mouse button and drag a box over a region containing peak crosses (without holding down any
keys). When the mouse button is released you will see that the peaks in the defined region are
highlighted with a border around the cross. To select just a single peak click near its centre with the left
mouse button. To delete the selected peaks press <Del> or select R:Peak:Delete selected.
If you have peaks selected and then select a different set of peaks you will see that the selection is
completely substituted for a new one. You can add to an existing peak selection by holding <Shift>
while you choose. Note that you can select peaks in several different spectra, and from different
windows, in this manner.
Markers and Navigation
Pick and select an isolated peak in the HSQC spectrum. Put a mark through it by holding the cursor
over the peak centre and pressing <m>. The lines produced are a multi-dimensional marker at the peak
position and will be visible at the equivalent 1H-15N location in the 3D window. To go to this
equivalent position in window2, with the cursor over the marked peak select R:Navigate:1H - 15N in
window2. Note that there are also navigation options for window3. There is only one navigation option
for window2 because it is an HCN window and there is only one way for the HN axes from window1
to map to it. However window3 has HHN axes and thus has two navigation options, where the second
option would take you to the 1H position on the vertical axis, rather than the horizontal axis, which
represents the amide proton.
Multi-dimensional marks, vertical ruler lines and horizontal ruler lines can be added to any window
location, not just on peaks, using the <m>, <v> and <h> keys respectively. To increase the number of
marks and rulers that can be displayed at one time select M:Window:Marks and Rulers or
R:Markers:Options. Note that you can clear all marks and rulers with the <n> key.
Strips and Strip Navigation
Now we will start to manipulate "strips" which are sub-divisions of a window that are connected (in
terms of their view) in one of the screen dimensions, but independent in the other dimensions. Go to
window2, select the "Strips" option at the top and click [+], this will add a vertical division to the
window. Click and drag with the middle mouse button to move the spectra - you will see that the
vertical axes of the strips are tied together, but the horizontal axis is independent.
The depth dimensions are also independent, e.g. if you move the bottom most, 15N scrollbar. Which
depth dimensions are moved depends upon which strip is active. The active strip is indicated by an
asterisk "*" next to its strip number and is set either by double-clicking (left mouse) within a strip or by
using the strip options at the top of the window; by clicking [1] or [2] or whatever. The active strip is
also the one that will be removed when the [-] button is clicked. You can rearrange the strip order,
moving the selected strip with the green arrows, and swap between vertical and horizontal strips with
the toggle button. To remove all the strips press the buttons with vertical and/or horizontal stripes to the
right of "Clear:".
Manually locating strips at interesting positions can be tedious, but there are various options to build
strips from pre-defined locations, for example using peaks. To make strips using peak locations select
three (picked) peaks in the HSQC spectrum and select (using the right mouse button) R:Strip:Peak
Location Strips:1H-15N in window2 (the same option is found in R:Navigate). The result is three strips
located at the amide positions corresponding to the HSQC peaks.
We are now done working with this project. You can quit Analysis via M:Project:Quit. It first asks if
you want to quit, and if you say Yes to that then it asks if you want to save the project, to which you
can reply No (unless you want to save what you have done).
Program Organization
Analysis stores the complete set of all relevant data in a set of files. Data are only saved when you
press save, so you can always quit without problems. There is an optional automatic backup system.
You can always kill a popup or window without causing any catastrophes.
Now we have seen some basic operations, we can look at how commands are organized. As you see,
things can often be done in more than one way, popups can be reached from several places, and
parameters - for peak picking, display, spectrum loading - are set automatically to sensible values. Still,
everything can be viewed and modified in detail. The top menu mirrors the conceptual organization of
the program. For instance things to do with spectra are under M:Experiment and things to do with
display under M:Window. For some things you might want to look in both places.
In any menu, options marked by a spanner are for viewing and editing the underlying data. Try looking
at the following, both for an overview and for some popups we have already seen:
M:Experiment:Spectra:{Spectra}
M:Experiment:Spectra:{Referencing}
M:Experiment:Experiments:{Experiments}
M:Experiment:Experiments:{Experiment Types}
M:Window:Windows:{Windows & Axes}
Popups for setting options are shown by crossed tools. Try looking at M:Peak:Peak Finding and
M:Peak:Draw Parameters to see what you can do with peak handling. General program options are
under M:Project:Preferences.
The other main types of popups can be seen on the M:Assignment menu. Tools are shown with
cogwheels - grey ones are lightweight or general purpose, while blue ones are complex special-purpose
operations. The colored squares icon is for data display and graphs. We will be getting to some of these
later.
The right mouse button in the graphics window has its own set of options that work off the cursor
position or the currently selected peaks. This is often the best way to access operations that have to do
with peak assignment, navigation, and display.
Initialising HSQCs
If CcpNmr Analysis is not already open, start it up again on the command line by typing:
-> analysis
(This assumes that the CCPN bin/ directory is on your path, otherwise you will need to type the full
path or be in the bin/ directory.)
Open an existing project
In the Analysis menu bar select M:Project:Open Project. Select [Yes] if Analysis asks whether it can
close the current project and [No] if it asks to save the current project. Navigate to find and select the
CcpnCourse1b project (the directory initially will be in green and then when you have selected it in
deep purple). Then click [Open].
You might get a warning that various files have moved location. You might also get a dialogue
box with a list of spectra paths (because those also have moved location). If the paths are all in grey
then just click the "All Done!" button at the bottom. If any path is in red then Analysis cannot find the
corresponding spectrum data file, so either you need to tell Analysis where it is (by double clicking the
path cell and navigating to the correct location) or accept that that particular spectrum will not have its
contours displayed.
This project has three windows, "window1", "window2" and "window3". The first is a twodimensional window with an HSQC spectrum in it, and the other two are three-dimensional windows,
one with an HNCA and an HNcoCA, and the other with an HSQC-NOESY. The HSQC has been peak
picked but not the other spectra.
Assigning New Resonances
Given a project with some picked peaks we will start some assignment of those peaks to resonances.
Initially we will assign the peaks anonymously, that is to say we will link peaks to a resonance number,
but not say which atoms the resonances comes from. Initially such an assignment is not very useful, but
we can go on to link related peaks to the same resonance numbers. For example we can say that a
whole column of peaks in the HNCA and 3D N-NOESY and an HSQC peak are derived from the same
amide resonances. When we specify which atoms the resonances derive from then all of these linked
peaks will automatically be assigned to those atoms.
To start an assignment choose the isolated HSQC peak at the location 7.27, 121.7 (1H, 15N), and with
the cursor near its centre press <a> (or you can select R:Assign:Assign HQSC...). Note it doesn't matter
which peak you choose really, but we will be referring to this one later. The Edit Assignment popup
will appear containing two rows of tables, one for each of the HSQC dimensions.
In the left most table of each row click the button [<New>], this will add the resonances [1] and [2] to
the 1H and 15N dimensions of the peak. Now click [Set Same Spin System] at the bottom left of the
popup. The resonance assignments will become {1}[1] and {1}[2] here the {1} annotation signifies
that the resonances are both in spin system number 1, which in this instance indicates that they both
belong in the same amino acid residue.
Now mark the peak (<m>), if it is not marked already, and navigate to the equivalent position in the 3D
N-NOESY spectrum in window2 via R:Navigate:1H - 15N in window3. Ensure the peaks along the
marker in window2 are picked (<Shift> + <Ctrl> + left click and drag). Note that if you cannot pick
some 3D peaks they may not have a maximum within the selected depth range. If this is the case then
you can adjust the depth position or width.
Now assign one of the 3D peaks at the marked amide location: Press <a> with the cursor over the peak
and find the Edit Assignment popup. You will see that the popup has now updated for the 3D peak and
consequently there are three dimension rows. Because the peak position closely matches the chemical
shift value for resonances [1] and [2] they appear in the right hand tables.
We can link these existing resonances to the 3D peak by clicking on their rows in the right-hand table.
When you do this the resonance annotation appears on the left hand side (and in the spectrum window)
to indicate that the peak is now linked, i.e. assigned to the resonances.
Note that you can remove the resonance assignment in the popup by selecting a row from the left hand
side and clicking [Clear Dim Contrib]. The use of the term "Dim Contrib" here reflects the fact that a
given peak dimension could potentially have a contribution from multiple resonances. So for example
you could have an ambiguously assigned NOESY peak where two different pairs of close resonances
contribute to the measured peak intensity.
With the 3D peak's amide dimensions fully linked we will quickly give the other 3D peaks at the same
amide position the same assignment. Do this by zooming out in the window so that you can see all the
peaks along the marker line, select all the peaks including the assigned one (left click & drag) then
select R:Assign:Propagate assignments. This will cause the resonances assignments {1}[1] & {1}[2]
(displayed on the spectra as "{1}[1],-,[2]") to be spread appropriately across all the selected peaks.
Efficient Resonance Assignment
The linking together of related peaks in different spectra by assigning them to common (anonymous)
resonances is something that can be partially automated to speed up the assignment process. Of course
you can also do this manually, as we illustrate above, if you wish. We can use the HSQC positions to
define unique amide locations and pick and assign related spectra based upon these "root" locations.
The first step in this automation is to define new amide resonance and spin system identifiers for all the
peaks within the HSQC spectrum. Select M:Assignment:Initialise Root Resonances. At the top left of
the dialog select the HSQC:115:1 Peak List if it is not already selected.
When you see the 'Initialise Root Resonances' popup, there is a table called 'Amide Sidechain Peaks'
with a few rows filled. Some of the peaks in the HSQC will be from NH2 groups of amine side chains,
and you need to handle those before you can initialise the peak list. Clearly the NH2 groups give two
peaks, one for each hydrogen, but both have the same 15N resonance (and thus 15N chemical shift). If
such pairs of peaks were processed in the same manner as the backbone amide peaks they would
become linked to two different pairs of resonances in two spin systems, when in reality they should
carry the same 15N resonance and be in only one (side chain) spin system. Click on a row of the table
to view the peaks, first making sure you set it to follow the right window (here "window 1"). If you
think this looks like side chain NH peaks, double click the 'Confirmed' column so it changes to 'Yes'.
Hint: there are five NH2 side chains in this protein.
When you are happy with all of them, click 'Initialise Peak List!' at the top.
This command calls the anonymous resonance and spin system assignment routines on all of the HSQC
peaks. Note that the routine knows which spectrum to work with because the experiment type of the
HSQC is set correctly as H[N]. You will now see that all of the peaks carry assignments of the form
{x}[y],[z]. If you look at the NH2 peaks that you confirmed, you will see that both peaks belong to the
same spin system and that the 15N dimension is assigned to the same Resonance.
The next part of the assignment process is to link resonances from the HSQC to the corresponding
trains of peaks in the 3D experiments. From the menu select M:Assignment:Pick & Assign From
Roots. In the Pick & Assign From Roots popup that appears ensure that window1 is selected in the
Root Window pulldown menu and select window2 in the pulldown menu in the Target Windows
section, and click [Add Target Window:]. Repeat this again to add window3 to the target list: select
window3 from the pulldown list and [Add Target Window] again.
Now take a quick look at the {Tolerances} tab and set the "Root 1H Dim1" tolerance to 0.03 ppm and
the "Root 15N Dim 2" to 0.2 ppm. Check if the other parameters look OK.
Then go on to the {Link Peaks} tab. You will notice that the peaks from the HSQC are listed in the
table. If you click on one of the rows, window1 will centre on that peak and the location of window2
and window3 will move to the same amide frequencies.
Rearrange the positions of the windows so that you can clearly see all of them, and the popup. Select a
row corresponding to an HSQC peak that is not overlapped and click [Pick & Assign Root
Resonances]. You will see that peaks are picked in the 3D spectra, in the box defined by the tolerances,
and assigned to the amide resonances from the HSQC.
You can go on to further amide positions by clicking [Next Root]. With appropriately set tolerances
you may also click [Pick All & Assign Root Resonances] to process all of the amide positions - you
can still use the 'Next Root' function (etc.) to loop through the peaks afterwards. Note that closely
overlapping amide resonances would still have to be checked or linked by hand.
It is important when picking peaks and assigning resonances in this automated manner that noise and
artefact peaks are not picked. Of course any offending peaks can be deleted afterwards, but most can be
avoided by setting the picking tolerances to appropriately small values and setting the contour levels so
that the noise is not visible. By default the peaks are picked only above the visible contour base level.
Now that we have defined and linked many resonances, look in the main resonance table at
M:Resonance:Resonances.
You will see that all of the resonances are listed here and many operations can be performed on them.
The important thing to note here is that the chemical shift of each resonance is automatically calculated
from the positions of the peaks to which it is assigned. Note that a resonance assigned to only one peak
will have no deviation in its shift, but those assigned to several 3D peaks will deviate as the amide peak
position varies slightly. By default the chemical shift values are the average of the assigned peak
positions, where each spectrum is weighted equally. However, different dimensions of different spectra
can carry different shift weightings (set at M:Experiment:Spectra {Tolerances}) so that the value of a
shift may be influenced more by the more precise experiments.
Data display tables
The standard data tables, like the Resonance table, have some useful features built in. Shift-click and
Ctrl-click allows you to select sets of rows. You can sort on individual columns by clicking the column
header. Clicking the right mouse button brings up a menu that lets you filter rows on their contents,
graph one column against another, and export sets of columns in a tab-separated format for scripts (or
Excel).
Entering sequences and molecular information
So far we have been linking related spectra with anonymous resonances. In the end we will assign
these resonances to specific atoms, but first we must specify the molecules within our experimental
sample that we may assign to. Creating a list of atoms to which we may assign has two distinct steps.
The first is the specification of a molecule, usually with an amino acid sequence, that will act as a
template. The second step is to build an assignable molecular system from the templates to give the
atoms. The molecular system represents all the assignable molecules that may reside within the NMR
sample, these include proteins, nucleic acids and small molecules. For this tutorial we make a mock
molecular system with one protein chain and one small molecule. These two steps exist so that we may
define a template sequence once, but have the potential to create several distinct polypeptide chains for
assignment, for example if we have a homodimer.
Open M:Molecule:Add Sequence. This will open the 'Molecule : Molecules' popup on the {Add
Sequence} tab. 'Add Sequence' is a shortcut command that can create both the molecule and molecular
system and add a new sequence to it, all in a single operation (it can also add to existing molecules and
molecular systems).
The kind of molecule you want to create is controlled by the switches at the top. First set 'Input Type'
to 3-Letter/Ccp. Then cut and paste the following sequence (on Linux: left mouse to select in the
browser and middle mouse to paste; on Windows: <Ctrl-C> / <Ctrl-V>; on OSX: <cmd-C> / <cmd-V>
into the Add Sequence text window:
LYS
MET
LEU
ASP
ALA
PRO
ASN
THR
SER SER PRO SER SER LEU THR TYR LYS GLU MET ILE LEU LYS SER
GLN
ASP GLY LYS GLY SER SER ARG ILE VAL LEU LYS LYS TYR VAL LYS
TYR
The input sequence does not have to be perfectly formatted. You can click [Tidy] at any time to see
how Analysis has interpreted the sequence.
Also, you can switch between one- and three-letter codes after the sequence has been entered. If you
look in the Ccp Codes pull-down menu (top right) you will see all of the residue codes that are
currently available, many of which are modified amino acids. Now set both 'Destination Molecule' and
'Destination Mol System' to <New>, and click 'Add Sequence!'. You will be prompted for the names of
the new molecule, molecular system, and chain. When finished the popup automatically switches over
to the {Chains} tab, where you can see the result.
Just for fun, we will now add a small molecule to our project. This could be part of the first molecule
we made, i.e. linked to the protein in some way, but instead we will make a new molecule. 'Add
Sequence' is only relevant for linear polymers so we shall do it another way. First go to the {Small
Compounds} tab. Begin by choosing [DNA] in the 'Mol Type' pulldown at the top then select a blue
highlighted compound in the right hand table to display an idealised structure in the left hand panel.
You can rotate the cartoon compound by clicking and dragging with the middle mouse button. Using
<Shift> or mouse wheel while dragging up and down with the middle mouse button will change the
zoom level.
With any compound selected set 'Destination Molecule' to '<New>' and click [Add Compound] to enter
it into your second molecule template - this will ask for a molecule name and then bring you to the
{Sequences} tab.
Go back to the {Small Compounds} tab and now try adding a non-polymer molecule. Set the 'Mol
Type' pulldown to 'Other' and select a molecule at random. Some simple molecules (like ATP and Zn)
are available, but most will give you a popup asking if it is OK to download the molecule description.
If you say 'Yes' the description will be downloaded (assuming you have an internet connection and
write access to the relevant directory), but even if successful most likely you will also get a warning
that there are no coordinates available (that is why we started with DNA). So select something like
ATP or Zn. Add that compound to another new molecule.
To make molecular system information from your templates, go to the {Chains} tab. Set 'Mol System
for new chain' to '<New>', select one of your small molecules as the template, and click [Make Chain
From Template]. Then set a new template, keeping the setting for the Mol System, and click [Make
Chain From Template] again. You are done creating molecules now; try to look in the various tabs and
see what information is available.
To view the new polymer chains have a look at M:Molecules:Atom Browser. You can scroll through
the sequence and by toggling the [H], [N] etc. you can display all of the assignable sets of atoms.
Triple-resonance protein sequence assignment
If the program is not already open, start CcpNmr Analysis on the command line by typing:
-> analysis
(This assumes that the CCPN bin/ directory is on your path, otherwise you will need to type the full
path or be in the bin/ directory.)
We will now look at the sequentially assigning protein backbone spin systems using triple-resonance
experiments. There are two basic, although linked, parts to the process. The first is the linking of
sequential spin systems (collections of resonances that relate to one residue) on the basis of matching
peak positions. The second is the matching of runs of unassigned spin systems to residues within a
sequence.
Open an existing project
We will now leave the previous CCPN project behind and load a new one; this differs from the old one
in that it has two more spectra; HNcoCaCB and HNCACB, and that all the peaks have been picked and
linked to the amide 'root'; resonances as described above (i.e. using the Assignment:Pick & Assign
from Roots option). In the Analysis menu bar select M:Project:Open Project. Select to close the exiting
project, but there is no need to save. Navigate to find and select the CcpnCourse2a project, then click
[Open].
Spectrum Setup
First make sure that window2 and window4 are visible and arranged such that window2 is tall and
narrow and on the left of a tall and wide window4 (both windows should be as tall as possible). Both
window2 and window4 have HCN axis and can display the triple resonance spectra. Note that to make
a new window you could use either use M:Window:New window or clone an existing window via
R:Window:Clone. Using the "Spectra" tab in the windows ensure that the HNcoCA and HNcoCACB
are the only spectra turned on in the narrow window and that all four triple resonance spectra are turned
on in the wide window.
Select M:Assignment:Protein Sequence Assignment. You should start out in the 'Window & Spectra'
tab. Select window2 as the 13C Window in the Query section and window4 as 13C Window in the
Match section. Make sure that the "Use" column is set to "Yes" for the query HNcoCA and
HNcoCACB spectra in the top table by double-clicking. For the bottom table set the "Use" column to
"Yes" for only the HNCA and HNCACB spectra (i.e. not the through-carbonyl experiments). This
setup means that we are going to compare specified 13C peak positions in the HNcoCA and
HNcoCACB experiments with potentially matching peak positions in the HNCA and HNCACB
experiments.
The rationale here is that the through-carbonyl experiment's peaks have the carbon shift of the
preceding alpha and beta carbons along the polypeptide chain at a given amide location, where the
HNCA & HNCACB have both the intra-residue and preceding alpha & beta carbon peaks for each
amide. Thus we can potentially use both spectra to say two amide spin systems are sequentially
connected; by saying that an inter-residue peak of the HNcoCA or HNcoCACB derives from the same
resonance as an intra-residue peak of the HNCA or HNCACB.
Note that this system can readily use other backbone experiments like HNcaCO, HNCO, HAcacoNH,
HAcaNH etc. with the same approach and that this tutorial only uses the alpha & beta carbon
experiments for simplicity.
Linking Sequential Spin Systems
Now go to the {Spin System Table} tab and in the to table scroll down and click on the row that
corresponds to spin system 8. Enlarging the popup window to the full height of the screen will help
view the tables more clearly.
You will see that the two triple resonance windows move to new locations. The location of window2 is
at spin system {8} and split into two regions one for the CA and the other for the CB peak. The other,
window4, has moved to the position of any peaks that match the carbon frequencies of the query peaks.
In this case there are three potential matches, but the first strip that corresponds to spin system {71} is
the only one that matches both the CA and CB positions well. Note that if the "Filter 13C By Inter/Intra
Type" option in the {Options} tab is set to off then spin system {8} matches even more strips. Because
we already have a good match we don't really need this option on, but it is useful if the previousresidue and same-residue peaks overlap significantly in the match strips. In the case of spin system {8}
matching {71} the CA position does not intersect the purple HNcoCA position, it clearly matches the
separate orange HNCA peak (and similarly for the CB position).
In the {Spin System Table} tab, go to the middle "Match Peak Positions" table a click to highlight the
row corresponding to spin system {71} (rank 1), and then click [Set Seq Link]. You will now see that
the tables of the popup update to show that {71} is set as "i-1" of {8}. Also note that in the spectrum
window the peak annotations of the aligned CA & CB peaks have changed to illustrate that they are
both assigned to the same 13C resonances.
Now click [Goto i-1], which will repeat the carbon shift matching, but this time from a sequence
position one earlier in the sequence. Repeat the above procedure for spin system {71}: select the best
match and link. For this exercise go on to sequentially link six spin systems in total. If all goes
according to plan, the order of spin systems (going i-1each time) will be {8} - {71} - {42} - {37} {48} - {41}.
Assigning Segments to the Sequence
You will see that as you select the various spin systems in the main table, in the lower right Residue
Types table there is a display of the probable types of amino acid residue. This prediction is based
upon how well the shifts within the spin system match the chemical shifts in the RefDB database. As
spin systems are connected sequentially, amino acid type predictions are made for the whole
sequentially connected section. In the lower left hand table the connected spin systems, given their
probable amino acid types, are matched to the protein sequence. Here the highest scoring positions of
residue type match for various five-residue sections are listed. You will see that the unassigned residue
positions are coloured grey, and the assigned regions become blue.
Click in the upper table on the row for spin system {37}, which is in the middle of our linked region,
and you will see that the residue type predictions are strong at and either side of this position. The
highest scoring option in the Sequence Locations table (hopefully with a score of about 66.7) should
correspond to the region from residue 17 Gly to 21 Leu, with the other sequence locations having lesser
scores.
Simply select the row for this highest-scoring location and click [Assign] and then [OK] to confirm the
assignment. You will see that all the residues in the section (by virtue of their links for the most part)
become assigned to the selected section, and that the colors in the 'Sequence Locations' table change.
Select the option M:Molecule: Atom Browser. Make sure that the elements [N] and [H] are displayed
(click the button to get the green hydrogen assignment options) and look at the amide atom for 19 Thr.
You will see that not only is 19 Thr assigned (i.e. the atom option goes dark green), but the atoms in
the residues which we just connected sequentially are also assigned.
Go through the spectra and the Protein Sequence Assignment (with 'Goto i+1') to verify that all the
connected spin systems have been assigned. Note that in this instance it was possible to assign the
resonances to unique atoms, as well as assign the backbone spin systems to the sequence, because the
resonances had their atom type set previously.
Automatic Protein Backbone Assignment
The semi-automated sequence assignment mechanism described above is supplemented in CcpNmr
Analysis by automatic assignment routines. At the moment only one called "Nexus" is publicly
available, but MARS, AutoAssign and other routines will be incorporated in the future. The automated
assignment routines are great for saving time, but they won't always work for all parts of proteins,
especially where peaks are missing or severely overlapped. In such instances you can run the
automation to assign the easy parts and then fill in the rest more carefully, where possible, using the
more manual routines.
Fortunately the data in the example CCPN project being used here is pretty good, and the peaks
positions have been checked and have undergone a degree of curation. Accordingly, we can assign
most of pure protein sequences automatically with a couple of clicks. To do this open
M:Assignment:Automated Seq. Assignment and in the resulting popup you will see that the four tripleresonance spectra are already selected; changing the "Use" column entry to "No" would mean the
spectrum was not used for the assignment.
All of these default settings are correct, so move on to the {Spin Systems} tab. In the spin systems
table you will be presented with all of the resonances, available in the chosen spectra, that are linked to
a backbone amide as either same-residue (Intra Residue) or previous-residue (Inter Residue). Initially
the table is quite bare, with only a few resonances (and their chemical shift values) displayed for the
few residues that we have already assigned. To fill in the rest of the table click [Find Resonances From
Peaks], selecting [OK] at the confirmation dialogue box. This function will then automatically extract
and label the CA and CB chemical shifts from the same- and previous-residue peaks. This is done by
considering the overlap of the through-carbonyl spectra with other spectra, together with the peak
intensities and relative chemical shifts that are expected to be visible (in an experiment of that type).
With the resonances populated in the spin systems table, move on to select the {Automation} tab. Here,
set "Use existing assignments?" to on and accept the other default settings.
Now click [Run Nexus]. Once the procedure has finished all of its iterations you will be shown a graph
of the assignment scores for each residue; where blue means good, yellow dubious and red bad.
Moving to the {Predictions} tab you will hopefully see large regions of good (blue) prediction.
Before a spin system (and thus all of its contained resonances) can be assigned to the predicted residue,
the prediction must be confirmed. To confirm the prediction either select a series of good rows (use
<Crtl> + click in the table) and click [Confirm Selected] or set all blue regions en masse by clicking
[Conform Above Score Threshold].
With your choices conformed click [Commit Assignments] and confirm [OK] to actually do the full
assignment; which will affect the peak labels.
Look at the peaks in window1 to see the result of the assignment, and admire your handywork.
Finding Side Chain Resonances Using TOCSY Data
The next kind of resonance assignment that we will look at is for the assignment of amino acid side
chain atoms. As an example we will choose a relatively simple residue, with relatively clear peaks,
naturally some residues with obscured or weak peaks will be more difficult to assign than this one, but
the general principles remain the same. Firstly we will load a new project with 3D TOCSY spectra
loaded. Select to close the exiting project, but do not save, then navigate to find and select the
CcpnCourse2b project and lastly click [Open]. This project contains an assigned HSQC spectrum,
some triple resonance backbone (HNC) experiments and three different kinds of three-dimensional
TOCSY spectra.
In window2 a three-dimensional CCcoNH TOCSY spectrum is shown. Like the HNcoCA and
HNcoCACB spectra, this shows peaks belonging to amide resonances that correlate with the carbon
resonances of the previous residue. For example, below we will look for the 20 Ala H,N (amide)
position but the side chain peaks on the Y-axis will be for residue 19 Thr.
In window3 a three-dimensional 15N HSQC TOCSY is shown. This spectrum shows side chain
hydrogen resonances on the Y-axis and amide (HSQC) resonances on the other axes. Here the amide
resonances are from the same residue as the amides.
In window5 a three-dimensional HCcH TOCSY is shown. This spectrum has two hydrogen axes and
one carbon axis. The hydrogen dimension on the X-axis represents resonances that are directly bound
to the carbon resonances on the Z-axis (depth axis). This contracts with the other side chain spectra
where the Z axis is for amide nitrogen. Thus here we are looking at CH pairs not NH pairs. The Y-axis
of this spectrum represents hydrogen resonances that are in the same residue as the bound CH pair on
the other axes.
In this example we will assign the side chain peaks for the 19 Thr residue, thus we expect to find three
carbon resonances; CA, CB, & CG2 and three hydrogen resonances; HA, HB, HG2* (methyl). To find
the hydrogen and carbon resonances in the amide-linked spectra we will navigate from the HSQC peak
positions. Accordingly, open M:Peak:Peak Lists, select the {Peak Table} tab and set the peak list
pulldown menu to "HSQC:115:1". Now sort the HSQC peaks by sequence by clicking on one of the
"Assign" column headings in the table. Scroll down so that peaks for 19Thr and 20Ala are visible.
Select the 20Ala row, then above the table set pulldown menu next to "Go To Position" to "1H - 15N in
window2" and click [Go To Position]. This will cause the display of window2 to move to the amide of
20Ala, but because the CCcoNH spectrum shows side chain resonances for the previous residue, the
peaks will derive from 19Thr in their middle dimension. Go back to the peak list table and select the
row for 19Thr, change the pulldown menu next to "Go To Position" to "1H - 15N in window3" and
click [Go To Position]. This will show the amide and side chain peaks for 19Thr in the HSQC TOCSY
spectrum.
With window2 and window3 located at the carbon and hydrogen side chain positions for 19Thr we
next assign the non-amide (middle) dimension of all the relevant peaks. For the CCcoNH spectrum in
window2 place the mouse over the peak labelled as "20Ala H,-,N" near 69 ppm on the Y-axis (carbon)
and press <a> to open the Assignment Panel. This is the CB peak for 19Thr, so when the Assignment
Panel opens, you will see "19ThrCb" as an assignment option in the middle table. Click on the
"19ThrCb" row to assign this to the middle dimension of the peak. Note that this CB resonance exists
and has a known chemical shift because if the assignments in the HNcoCACB and HNCACB
backbone experiments.
Repeat this procedure for the peak at 66 ppm ( press <a> over it) to give it a "19ThrCa" assignment in
its middle dimension.
Assigning New Side Chain Resonances
For the last carbon position at 20 ppm we will make an assignment to the CG2 atom, however this
resonance is new to the project, and hence there is no assignment suggestion that appears automatically
for the middle dimension. Instead we will add a new resonance that we will link to the right atoms. To
do this click on the middle [<New>] button (F2 dimension). You will see a new resonance number
appear of the form [number] in the middle dimension. Click on this new resonance in the left most
column, then click [Assign [number]] at the left of the blue buttons. In the Atom Browser that appears,
switch on the hydrogen and carbon atoms using the[C] and [H] buttons. Then scroll down to the 19 Thr
residue and click on the right most "Cg2" (the "g" will be a Greek gamma). Returning to the
Assignment Panel you will now see that the resonance assigned to the middle dimension is labelled as
"19ThrCg2".
Next move to window3 and assign the side chain peaks of the HSQC TOCSY spectrum in a similar
way. In this case the peaks you need to consider will be initially labelled as "19Thr H,-,N" because the
middle dimension will carry the hydrogen assignments for the same residue as the amide. There will be
three peaks at 4.26, 3.82 and 1.21 ppm, these correspond to the HB, HA and HG2 resonances
respectively. Note that the HB peak is below the HA peak because this is a Thr residue. For each of
these peaks you will need to make a new resonance with [<New>] and use [Assign [number]] to link it
to the right atoms.
Assigning the HCcH TOCSY
For the HCcH TOCSY spectrum we will use the location of the side chain resonances we have just
assigned in the other TOCSY spectra to locate peak positions. Note that this spectrum does not have
any peaks picked in it. The idea is that the CCcoNH spectrum will provide the ppm positions for the
carbon resonances (on the Z-axis) and the HSQC TOCSY will provide the ppm positions for the
hydrogen resonances (X- and Y-axes). The lines of intersection between the carbon and hydrogen
resonances will hopefully show where the peaks (with HHC dimensions) can be found within the
HCcH spectrum.
Open window5 to view the HCcH TOCSY spectrum. You can close the other windows to save space.
Go to the main Analysis menu and select M:Resonance:Spin Systems. In the Spin Systems table that
opens, click on the "Residue" column to sort the spin systems (groups of resonances in a single residue)
into sequential order. Scroll down to the row for 19Thr, select the row, check that window5 is selected
in the top right pulldown menu, and click [Display Strips].
In window5 you will hopefully see three strips; for the CA-HA, CB-HB & CG2-HG2* positions.
With 19Thr and window5 still selected in the Spin Systems table click [Display Cells]. You will now
see that the display in window 5 is now also split vertically to give separate HG2*, HA, HB and HN
regions: these are for all hydrogens in the spin system although the HN resonance is not visible in the
HCcH TOCSY, however it would be in a 13C HSQC NOESY.
For each of the nine intersection points with obvious maxima pick new peaks (<Shift> + <Ctrl> + left
click). Assign each peak by pressing <a> with the mouse over the peak cross. The relevant resonances
should be automatically presented for you, so all you have to do is click on the options in the right hand
side of the Assignment Panel. Note that you can assign several the peaks that align vertically or
horizontally to the same resonances (in the matching dimensions) by selecting the peaks (left <Shift> +
click) and spreading the assignments with R:Assign:Propagate assignments. Note that the 19Thr Ha,
Hb, Ca peak at 3.82, 4.26, 66.0 ppm is slightly distorted along the Z-axis, so you may have to manually
place the peak in the CA plane before it can be assigned to 19ThrCa; either pick it with <Ctrl> + click
or select it and press <p> while in the exact CA plane. You might also have to click “Double
Tolerances” in the Assignment Panel to see all the matching resonances.
Copying Assignments, Multiple Shift Lists
If the program is not already open, start CcpNmr Analysis on the command line by typing:
-> analysis
(This assumes that the CCPN bin/ directory is on your path, otherwise you will need to type the full
path or be in the bin/ directory.)
Setting up and linking a new shift list
So far during this tutorial, all of the chemical shift values we have recorded, by picking peaks and
linking resonances & atoms, have gone into one single list. However, there are several circumstances
when it is helpful to use more than one list of chemical shifts, so that you may for example separate
sets of shifts based upon the experimental conditions they relate to; you may use different shift lists for
different temperatures or when doing titration experiments. Commonly in such circumstances the
underlying resonances (and hence atoms) that we work with are the same, but the positions of the peaks
move significantly and we would not want to average chemical shift values over such a variation.
In the Analysis menu bar select M:Project:Open Project. Select to close the exiting project, but there is
no need to save. Navigate to find and select the CcpnCourse2c project, then click [Open].
We are going to set up a new shift list for the second HSQC experiment present in the demonstration
project. This second HSQC experiment relates to exactly the same sample (molecule) as the first
HSQC but was recorded at a different temperature, which has caused the peak positions to move.
Given that the second HSQC experiment is already loaded, all we need to do is go to the
M:Experiment:Experiments {Experiments} and in the Shift List column double click on the cell
corresponding to HSQC_2. In the pulldown menu that appears simply select <New>. You will see that
a new shift list is now listed.
From now on whenever an assignment is made in HSQC_2 the shift remains separate from the other
spectra. Indeed, any resonances that were already connected to HSQC_2 before its shift list was
changed will have their shifts recalculated in two separate groups. You could move the experiment
back to the original shift list at any time and the chemical shift values will be appropriately
recalculated. However, if you disconnect experiments from a shift list (or delete a peak etc) you might
have chemical shift values that are not defined by any peaks. Under such circumstances the chemical
shifts persist, but are described as "orphans".
Assignment of HSQC_2, with its separately curated shifts proceeds pretty much as assignment
normally does, with the only major difference being that for a new shift list (i.e. without any
assignments under it) there will be no resonance possibilities that can appear when assigning a peak.
The only known shifts are distinctly different, representing different conditions.
To assign HSQC_2, even though we don't have shifts set, we can say that certain peaks in the two
HSQC spectra are equivalent if we can see how they have moved under the different situations.
Accordingly we can select a peak in the first HSQC and the corresponding peak in HSQC_2 and
propagate assignments. That is to say that the peak dimensions link to the same resonances (and hence
atoms), even though things have moved. Go to spectrum window1 and try this for any pair of close,
isolated peaks; one peak in each of the HSQC spectra. Use the mouse with a left click to ensure that
these peaks (and only these peaks) are selected. Then in the right mouse menu select
R:Assign:Propagate assignments. Note that this function usually checks to ensure resonance positions
are within tolerances, but here with the two shift lists it cannot so the connection is made anyway,
which is exactly what we want.
You may like to repeat the above procedure for several HSQC peaks and their counterparts.
With a few resonances represented in the new shift list (should be "ShiftList 2") have a look at the
resonance table M:Resonance:Resonances table to see evidence of these links. In the resonance table
note that you can choose between the different shift lists in a pulldown menu at the top, i.e. set the
ShiftList: pulldown to "ShiftList 2". Note that when the shift list changes the resonance positions shift
and the number of peaks linked also changes.
Apart from the Resonance Table, Analysis also provides direct access to the shift measurements.
Looking in the menu at M:Data Analysis:Measurement Lists, you will see that there are tables of the
shift values corresponding to each list. Note that other types of value like T1 rates, or Hetero-NOE
values will also appear as measurements in these tables if they are calculated.
Copying peaks between related spectra
Propagating assignments between peaks by manually selecting them, although sometimes necessary,
can be tedious if you have large number of peaks that are equivalent and have closely matching
positions. In this instance Analysis provides a utility to compare all of the peak positions in two related
spectra and transfer the resonance assignments efficiently, whilst still leaving sufficient scope for
human intervention.
In this exercise we will transfer the amide assignments from the peaks in the HSQC experiment to the
HSQC_2 experiment. In the main menu we select M:Assignment:Copy Assignments. In the Copy
Assignments popup ensure that the first "HQSC:115:1" experiment is set as the Source Peak List and
that the second "HQSC_2:48:1" is set as the Target Peak List. In the top table you will see a list of the
source peaks, together with an indication of how many destination peaks they match, within the given
tolerances. Note that when the Follow Peaks? option is set clicking on a source row will cause peak to
be highlighted in a relevant window (window1). Also, selecting the source will display the potentially
matching destination peaks in the lower table. Clicking on a target row will, as before, highlight the
relevant peak.
This utility can be used in one of two ways. One can manually choose the correct destination for each
source peak and [Assign Selected Target], or if you trust the completeness and accuracy of the
destination one can automatically assign the closest destination peak to each source one with [Assign
Singly Matched]. When taking the more automated approach it is common to start with strict tolerances
to assign the more certain matches and then relax the tolerances to fill in the remainder.
Relaxation Rate Analyses
The CCPN project CcpnCourse2c contains three spectra used to calculate NMR relaxation rates, plus
an assigned HSQC spectrum than can be used as a reference. The rates analysis spectra are represented
as pseudo-three-dimensional data sets; where different 2D spectrum planes that correspond to various
T1 and T2 delay times have been stacked atop one another to give a 3D spectrum. Within this type of
data set the third dimension is not in PPM, it is delay time and corresponds to the "z" axis - i.e.
orthogonal to the X-Y plane that is the screen. The procedure used to analyse the data is very similar to
the analysis of chemical shift titrations; where we follow the position of peaks, rather than the
intensities of peaks. However, to keep this tutorial simple we focus on only the relaxation analysis.
Relaxation Analysis
There are three experiment series loaded into this project; and each corresponds to a separate pseudo3D experiment (although it would be possible to use a series of 2D spectra). There are two T2 series
and one T1 series, the basic analysis of which involves plotting peak intensity verses delay time for
each clear amide position, and then fitting the intensity curve with an exponential function to extract
the T1 or T2 parameter; which is deduced from the rate of exponential decay.
Firstly look at the T1 rates experiment by selecting window1. Switch off the HQSC and put on the T1
experiment. Note at the bottom of this window is a different kind of scrollbar, made up of buttons.
These buttons control which planes of the pseudo-3D experiment are visible on screen.
With the left mouse button you can shift the visible planes through the series, and with the middle
mouse button (try both left and right buttons together if you don't have a middle button) you can extend
the number of visible planes. Try this and switch on all of the planes of the experiment.
If you pick peaks here, you will be picking peak maxima in all planes that are visible. If you don't want
pick in a specific plane, simply ensure that plane is switched off. We could pick and assign all the
peaks in the T1 series, however, if you toggle on the [HSQC:115] spectrum (use the [Spectra] option at
the top of the window), you will see that this assigned HSQC aligns well with the T1 series. We will
use this fact to pick peaks and to spread the HSQC assignments to all of the planes of the T1
experiment when we do the relaxation analysis.
Before we do the relaxation analysis go to M:Experiment:NMR Series. Clicking on the "T1" row you
will note that the time values for the series are already entered. This is because the spectrum was
loaded from a parameter file (AZARA format in this instance) where the time points of the series are
already specified. You could still change then here if you wished. Anyhow, in the top table note that
the "Condition Type" is set to "delay time", but it can be changed as required. Before you close the
popup make sure that 'Unit' is set to seconds ('s').
Now select M:Data Analysis:Follow Intensity Changes. You will note that an Experiment Series is
already selected, by virtue of it having the "delay time" as the parameter type. However, it is probably
not the correct one in this instance, so ensure the "T1:delay time" option is selected.
Ensure that the Reference Peak List is set to "HSQC:115:2" this is a peak list we have assignments for
and has any troublesome weak or overlapped positions removed. Set the Fitting Function to "A exp(Bx)"; i.e. an exponential decay. Set the 1H tolerance to 0.05 and 15N tolerance to 0.1 - this indicates
the size of the region with which analysis will try to find or pick peaks in the pseudo-3D experiment,
based on the locations in the assigned HSQC.
Then click [Group Peaks]. If some fits fail just click 'OK'.
You will see that the peaks have been picked and that the assignment has propagated across the whole
series and that the selected function/graph has been fitted to the peaks.
Selecting an assignment and clicking on [Show Fit Graph] and [Next Set] allows you to quickly
investigate the quality of fit to the intensity for all peaks in each group.
Note that the time constant for each group is recorded in the "Time Constant" column. This is
calculated from the inverse of the exponential decay rate; i.e. the "B" in "A exp(-Bx)". You may draw a
graph for the T1 values or export the data as text, and you can choose to create a dedicated T1 list by
pressing [Make T1 List] - make sure you do this for the next stage. This simply records the T1 values
(as indicated by the "Time Constant" column) in a list of NMR measurements that are saved when your
CCPN project is saved. Accordingly this data may be examined at any time in the future (and even
deleted) by selecting M:Data Analysis:Measurement Lists and selecting the T1 row then [Show
Measurements].
T1 vs T2 Macro
Finally, we will look at loading an external module to analyse the relaxation data further. We will be
using the "Macro" system in Analysis, which is a means of importing and running modules that
manipulate the CCPN data using Python scripts. Here we will be loading in a Python script that plots
the newly calculated T1 values against the T2 values so that we might have some idea about the
relaxation properties of the molecule; in this instance we will get a visual clue about order parameters
and molecular correlation time. For this exercise it is perhaps more important that you understand the
idea of how Analysis can be expanded in this manner, rather than learn the specific details of this
module (which is actually a work-in-progress). If you want to know more about programming for
CCPN, see the examples at http://www.ccpn.ac.uk/ccpn/data-model/python-api-v2-examples/.
If you don't already have the Python script you can download it at
http://www.ccpn.ac.uk/ccpn/projects/relaxation-analysis/relaxationanalysis.py.
From within Analysis go to M:Macro:Organise Macros.
In the resulting popup click [Add Macro] at the bottom right corner. You will then be presented with a
file browser where you need to navigate to the location of the saved Python script
"relaxationanalysis.py". When you finally click on the row of the Python file in the top file table you
will notice that the lower table is filled in with all of the subroutines (functions) that are available
within the Python module. Select the first function in the table called "relaxationAnalysisMacro".
Then press [Load Macro] at the bottom. After a short pause the new module will be loaded and the
selected function will appear at the bottom of the Macros table; so you may need to scroll down to see
it. In the row for the new macro double-click in the "In main menu?" column so that it becomes a green
"yes" (or purple if highlighted), and then close the popup.
The newly loaded Python script is now available from inside Analysis via the main menu, i.e. at
M:Macro:relaxationAnalysisMacro.
Select this option and see what it does. (Note on some systems you may have to resize the popup to see
the plot).