Supplementary Methods. - Word file (61 KB )
advertisement

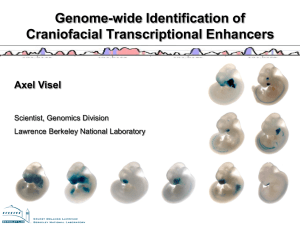
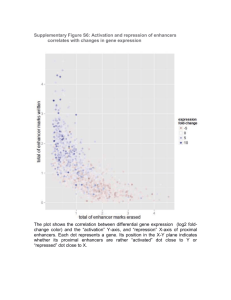
Supplementary Methods Identification of Conserved Noncoding Elements. The set of human-fugu conserved noncoding elements tested in vivo was derived from a computational pipeline described previously1. Using genomic DNA alignments, we constructed a syntenic map defining homology between the human and fugu genomes. We then identified discrete fragments showing conservation (70% identity with a match minus mismatch score2 ≥ 60). Transcribed sequences in the conserved set were filtered out using known genes, spliced ESTs and mRNA annotations obtained from the UCSC genome browser (intronic conservation was allowed). We then manually curated the data set to remove any additional false-positives by visual examination of UCSC genomic data. Whole-genome human-mouse-rat and human-mouse-fugu conserved noncoding elements were subsequently identified and assigned p-values using Gumby3 in combination with a more recent version of the program used to construct synteny maps4. In the course of our study, we initially focused on human chromosome 16 and tested 79 elements (73 were human-fugu conserved, 4 were human-fugu-ultra conserved, and 2 were ultraconserved alone) from this chromosome but subsequently expanded the study to include 88 elements on other chromosomes (10 were human-fugu conserved, 50 were human-fuguultra conserved, and 28 were ultraconserved alone). The primary rationale for this expansion was to compare the success rate of enhancer identification of human-fish versus human-ultra conservation. Mouse transgenic enhancer assay. Primers were designed to flank the conserved element by several hundred basepairs using primer35 and can be found for each element at http://enhancer.lbl.gov/. Enhancer element constructs were PCR amplified from human genomic DNA (BD Biosciences) and directionally cloned into the pENTR/DTOPO vector (Invitrogen). All inserts were sequence-validated and transferred into a Hsp68-LacZ vector6 encompassing a Gateway cassette using LR recombination (Invitrogen). Generation of transgenic mice and embryo staining was performed as previously described7 in accordance with protocols approved by the Lawrence Berkeley National Laboratory. Transgenic mouse DNA was prepared from yolk sacs that were carefully dissected from embryos, boiled for 5 minutes in lysis solution (50 mM Tris HCl pH 8.0, 20mM NaCl, 1mM EDTA pH 8.0, 1% SDS), and then screened by PCR with LacZ primers (LacZ-fwd 5'-TTTCCATGTTGCCACTCGC; LacZ-Rev 5'- AACGGCTTGCCGTTCAGCA) for positive transgenic animals. Images were obtained using a Leica MZ16 microscope and DC480 camera, cropped and level adjusted with Adobe Photoshop. High-resolution images of single embryos were deposited into our internal database. Positive enhancer scoring and annotation. For each enhancer fragment, all transgenic embryos exhibiting LacZ-staining were scored and annotated independently by multiple curators. Positive enhancers required a minimum of 3 independent transgenic embryos showing a consistent expression pattern (though 83% had 4 or more, and 67% had 5 or more supporting embryos) while negatives required no obvious consistent reporter gene expression and/or at least 3 non-staining mice that were also positive for the transgene as determined by PCR7. Nomenclature standards were obtained from Bard et al8 and, in general, used a low-resolution vocabulary based on whole embryo microscopy visualization. Length and Conservation of Positive versus Negative Enhancer Sequences. To investigate possible predictive features of positive enhancers, we mapped 160 of the 167 tested sequence elements to the whole-genome set of syntenic human-mouse-rat conserved noncoding elements3,4. We selected the human-rodent dataset to enable assessment of both the human-rodent-ultra- as well as human-fugu- tested fragments, with 7 of the assayed elements eliminated due to limited synteny or missing sequence in the rat genome. We found that positive enhancers overlapped human-rodent conserved elements with a mean length 1,630 bp, many of which extended beyond the boundaries of the tested sequence, whereas the negative enhancers mapped to significantly shorter (ttest p value=0.0087) human-rodent elements (mean: 966 bp). Similarly, the positive enhancers mapped to human-rodent elements with significantly higher (t-test p value: 0.0004) evolutionary conservation scores (mean Gumby -log(p value): 67.1) than the negatives (mean -log(p value): 43.5)4, indicating that the degree of conservation between humans and rodents can be used to further prioritize human-fish and ultra-conserved elements for functional activity under this experimental design. It is worth noting that a previous study9 also indicated that of 15 less conserved sequences tested in this assay, only one functioned as a developmental enhancer. Motif-finding in a preliminary set of human enhancers. To find sequence motifs that were associated with particular expression patterns, we used a discrete, enumerative motif-finding approach10. We focused our training set on all tested human-fugu elements from chromosome 16 which comprised our first available dataset, where 4 of the 77 tested fragments yielded strong forebrain enhancer activity. Because the training set was small (4 fragments, totaling 1,090 bp of conserved sequence), we chose to search for words of length 5 to retain statistical power. We tested all the 5-mers (allowing a spacer of up to 2 before the 3rd base) against the null hypothesis that they appeared in the 4 robust forebrain enhancers as frequently as in a background set (see below). We assigned n n significance using the binomial distribution P( x | n, f ) f k (1 f ) n k where P is kx k the probability of observing x or more given n tries and frequency in the background set, f. We chose a significance threshold of 1 , which we expect to produce less than 3 45 one motif by chance since we treat each 5-mer and its reverse compliment as one motif (there are only 2560 motifs tested) and the motifs are not truly independent. When we identified a 5-mer that exceeded our threshold, we removed it from the training set, and repeated the procedure until there were no 5-mers that exceeded the significance threshold. Using this procedure we searched for motifs that were enriched in the forebrain enhancers relative to three sets of background sequences: 1) random sequences from chromosome 16 (which yielded ATTAA and GATTA, which we note are motifs present in previously characterized embryonic forebrain enhancers11,12), 2) the chromosome 16 set of human-fugu fragments (which yielded TTNNAAA, CANNGGC and TANNTGA) and 3) the chromosome 16 set of human-fugu sequences that displayed enhancer activity (which yielded TTNNTTT). Because the latter two comparisons are between sets of sequences for which we have alignments with mouse and fugu, in those cases, rather than counting the 5-mers in the human sequence, we counted conserved 5mers (defined as a match in the same position in each species in the alignment of human, mouse and fugu). We note that motifs identified in each of these comparisons have slightly different interpretations, and we decided that all might be important for de novo forebrain enhancer prediction. Predicting forebrain enhancers. Because tissue-specific enhancers often contain multiple binding sites for multiple transcription factors, we sought to combine information from all the motifs for the prediction of new forebrain enhancers in the genome. We scored each of 3,124 human-mouse-fugu noncoding alignments for the number of conserved (found aligned in human-mouse-fugu) matches to each of the 6 significant 5-mers4. We ranked the fragments using a score that compares the frequency of conserved motif matches (as defined above) in the fragment to the expectation based on the background frequencies (f) over all the fragments. The score, S, for the ith fragment is given by Si mmotifs xmi log xmi ni , where the sum is over each of the motifs, fm m, x is the number of times a particular motif occurred in a particular fragment and n is the length of a fragment. The top 30 fragments are available in Supplementary Table 3. Assessing significance of predictions. We assessed whether our motif enrichment and conservation-based prediction method was more effective than using conservation alone by attempting to reject the null hypothesis that k1 successes out of n1 tests (in the training data) and k2 successes out of n2 tests (in the test set) were obtained from the same distribution. We calculated the probability of observing k2 or more successes out of n2 draws from a binomial distribution, integrating over all possible values of the binomial probability p weighted by the posterior probability of observing k1 successes out of n1 tests for each value of p (the integrals were estimated numerically): n 2 n2 n1 k1 k n1k 1 p (1 p) n 2k dp p ( 1 p ) 0 k1 k k 2 k P(k1, n1, k 2, n2) 1 n1 k1 n1k 1 0 k1 p (1 p) dp 1 Potential scoring bias. Because our motif conservation score is based on the number of conserved motifs, the top predictions tended to be more conserved and longer than the average. Since we had found that longer, more conserved fragments are more likely to function as enhancers in our assay, we considered the possibility that the enrichment of forebrain enhancers was simply due to an overall increase in enhancer prediction. We found, however, that the fraction of fragments that showed expression patterns other than forebrain in the chromosome 16 set 17/77 was not different to the fraction observed for that in the predictions 7/23 (p value=0.28), suggesting that the frequency of enhancer discovery for patterns other than forebrain had not changed. Supplementary Methods References 1. 2. 3. 4. 5. Grimwood, J. et al. The DNA sequence and biology of human chromosome 19. Nature 428, 529-35 (2004). Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520-62 (2002). Prabhakar, S. et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res (2006). Ahituv, N., Prabhakar, S., Poulin, F., Rubin, E. M. & Couronne, O. Mapping cisregulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet 14, 3057-63 (2005). Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132, 365-86 (2000). 6. 7. 8. 9. 10. 11. 12. Kothary, R. et al. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105, 707-14 (1989). Poulin, F. et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85, 774-81 (2005). Bard, J. L. et al. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev 74, 111-20 (1998). Nobrega, M. A., Zhu, Y., Plajzer-Frick, I., Afzal, V. & Rubin, E. M. Megabase deletions of gene deserts result in viable mice. Nature 431, 988-93 (2004). van Helden, J., Andre, B. & Collado-Vides, J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281, 827-42 (1998). Kurokawa, D. et al. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131, 3319-31 (2004). Zhou, J., Zwicker, J., Szymanski, P., Levine, M. & Tjian, R. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc Natl Acad Sci U S A 95, 13483-8 (1998).