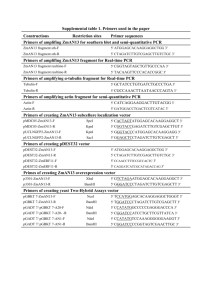
Plasmids
advertisement

Supporting Materials and Methods Plasmids pT/KRas-G12V was constructed by inserting a KRas-G12V fragment into the pT3/EF1α plasmid. The corresponding fragment was generated by PCR from the plasmid pORF9-mKRas2 (InvivoGen) using ATGACTGAGTATAAACTTGTGGTGGTTGGAGCT the primers G12V-fw: GTTG and G12V-rev: GTATTCACATAACTGTACACCTTG. pT/KRas-G12V-IRES-GFP was cloned by coupling IRES-GFP (pQCXIX, Clontech) with KRas-G12V into pT/KRas-G12V. pT/EF1α-EGFP was designed by cloning of EGFP into pT3/EF1α. The plasmid pT/mAlb-EGFP was constructed by replacing the EF1α-promoter in pT/EF1α-EGFP by the mouse albumin-promoter from the plasmid pAlb.GH (kindly provided by Mark A. Magnuson, Vanderbilt University). To construct pT/mCK19-EGFP, the murine CK19-promoter was amplified by PCR using GTTCCTTTCTAAGACCCACCAGTC the primers and CK19-fw: CK19-rev: GAGACCAGAGCGAAGGAAAAAG. The resulting fragment was inserted into pT/EF1α-EGFP thus replacing the EF1α-promoter. Detection of Cre-mediated Trp53Δ2-10-recombination in tumor cells Induced tumors were isolated, dissected, and incubated 30 min at 37 °C in RPMI1640+Glutamax (Life Technologies) supplemented with 200µg/ml of collagenase IA, collagenase IV, and hyaluronidase IV, 300µg/ml dispase, and 50µg/ml DNase I (all Sigma). Separated cells were filtered through a 40µm strainer, and cultured under standard conditions. After 8 passages, cells were counted and single cells were seeded in 96-well plates by limiting dilution. Growing colonies were analyzed for RT-qPCR or p53floxgenotyping. DNA was isolated using QIAmp DNA Mini Kit (Qiagen). Primers for detection of Trp53F2-10 alleles by PCR were described previously (1). For detection of the 5´loxP-site, intron 1 PCR yields a 370 bp or 288 bp fragments for the floxed or wildtype alleles, respectively and for the 3´loxP site, intron 10 PCR yields a 584 bp or 431 bp fragments for floxed or wildtype alleles, respectively. In case of recombination of Trp53Δ2-10 allele, 612-bp fragment can be observed. For detection of control exon 11, PCR yields a 390 bp fragment. Reverse Transcription qPCR Total RNA was isolated using the RNeasy-Mini-Kit (Qiagen). Random hexamer primer and TaqMan RT-Reagents (Applied Biosystems) were used to generate cDNA. For quantification, SYBR-Green (Eurogentec) and Lightcycler®480II (Roche) was used. Ct-values of amplicons were normalized by GAPDH as internal control. Primers for EGFP (2) and p53 (3) quantification were described previously. Following primers were used for mKRas and mGAPDH quantification: KRas-fw: GTAAAGGACTCTGAAGATGTGCC; KRas-rev: CAATGAACGGAATCCCGTAACTC; GAPDH-fw: AGAACATCATCCCTGCATCC; GAPDH-rev: CACATTGGGGGTAGGAACAC. References 1. Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van d, V, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 2001 Dec;29(4):418-425. 2. Lalancette C, Thibault C, Bachand I, Caron N, Bissonnette N. Transcriptome analysis of bull semen with extreme nonreturn rate: use of suppression-subtractive hybridization to identify functional markers for fertility. Biol Reprod 2008 Apr;78(4):618-635. 3. Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011 Mar;13(3):317-323.