Supporting Text Phylogenetic analysis of selected isolates: PCR
advertisement

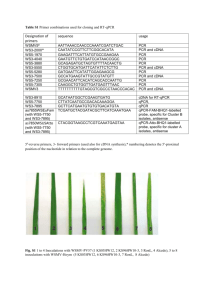
Supporting Text Phylogenetic analysis of selected isolates: PCR amplification of genes: Out of the 76 “C. capsici” isolates obtained from UAS, 8 isolates were selected for taxonomic identification based on multigene sequence analysis. The 8 isolates isolates were subjected to PCR amplification of the partial actin (act), chitin synthase (chs1), glyceraldehyde-3-phosphate dehydrogenase (gapdh) and histone (his3) genes. The cultures procured from MTCC and ITCC were subjected to internal transcribed spacer region (ITS)/ 5.8S rRNA sequencing. PCR was performed in a 50 µl reaction volume. Each reaction tube contained 3 µl of total genomic DNA, 10 µl of 5x FirePol™ PCR Mastermix Green Buffer (Solis Biodyne, Estonia), 1.5 µl each of 10 µM forward primer and reverse primer and 34 µl of sterile water. PCR reactions were performed in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) using cycling parameters and primers as previously described (Prihastuti et al. 2009 – ITS (ITS 4 and 5), act (ACT512F and ACT783R), gapdh (GDF1 and GDR1); Damm et al. 2009 – chs1 (CHS-79F and CHS-354R), his3 (H3F and H3R)). PCR products were run on a 1% Tris-Acetate-EDTA agarose gel stained with ethidium bromide (0.5 µg/ ml) at 100 V, 400 mA for 45 minutes to check the presence of desired band. The PCR products were purified using QIAquick PCR Purification Kit (QIAGEN, Catalogue number 28106, Hilden, Germany). The purified PCR products were quantified using Nanodrop Spectrophotometer ND-1000 (Thermo Fisher Scientific, Delaware, USA). The PCR products were sequenced using their respective primers with ABI Big Dye v3.1 Terminator Ready Reaction Cycle Sequencing Kit (Applied Biosystems) using manufacturer’s protocol. The samples were purified to remove excess salt and unincorporated dNTPs, denatured with HiDiFormamide at 95 ºC for 3 minutes and analyzed using 3730 DNA Analyzer (Applied Biosystems) at the central DNA sequencing facility at the Institute of Microbial Technology, Chandigarh, India. Multigene phylogenetic analysis: The forward and reverse sequences obtained for each strain were aligned using SEQUENCHER v.4.10.1 (Gene Codes Corp., Ann Arbor, Michigan, USA) to generate a consensus sequence. A concatenated multi-gene dataset comprising act, chs1, gapdh and his3 genes for the 8 “C. capsici” isolates from UASB and selected reference type sequences from Damm et al. 2009 was generated using SequenceMatrix v.1.7.8 (Vaidya et al. 2011). Maximum parsimony (MP) analysis of the multigene dataset was performed using PAUP version 4.0b10 (Swofford 2003). Ambiguously aligned regions were excluded from the analysis. The gaps in the alignment were treated as missing data. All the characters in the analysis were unordered and had equal weight. Trees were inferred using the heuristic search option with 20 random sequence additions and tree bisection and reconstruction (TBR) as the branch swapping algorithm. Maxtrees were set to 10,000; branches of zero length were collapsed and all multiple parsimonious trees were saved. Descriptive tree statistics [Tree Length (TL), Consistency Index (CI), Retention Index (RI), Related Consistency Index (RCI), Homoplasy Index (HI)] were calculated for the generated trees. The robustness of the trees was measured by 100 bootstrap replicates (Felsenstein 1985) and addition of 10 random sequences. Kishino-Hasewaga (KH) tests (Kishino and Hasewaga 1989) were performed in order to determine whether trees were significantly different. Trees were figured in Treeview (Page 1996) and edited in MEGA version 5.05 (Tamura et al. 2011) and Microsoft PowerPoint version 2007 (Microsoft Corp., USA). The sequences generated in this study have been deposited in NCBI-GenBank with accession numbers as listed in Supporting Table 1. The alignment files and trees are deposited in TreeBASE (www.treebase.org; Study ID: 14452). The resulting MP tree is shown in Supporting Figure 1. ITS- based phylogenetic analysis: 23 Colletotrichum isolates procured from MTCC and ITCC were selected for ITS based phylogenetic analysis. A maximum parsimony analysis of the ITS dataset for ITCC and MTCC isolates along with selected reference type sequences from Damm et al. 2009 was performed using PAUP version 4.0b10 (Swofford 2003) as already described above. The sequences generated in this study have been deposited in NCBI-GenBank with accession numbers as listed in Supporting Table 2. The alignment files and trees are deposited in TreeBase (www.treebase.org; Study ID: 14452). The resulting MP tree is shown in Supporting Figure 2. Results: Multigene phylogeny: There were a total of 1227 positions in the multigene dataset. The gene boundaries in the multigene dataset included: act: 1–254, chs1: 255–505, gapdh: 506–842 and his3: 843–1227. The analysis involved 42 nucleotide sequences. Hundred characters from the ambiguous regions were excluded from the analysis. Out of the remaining 1127 characters, 817 characters were constant, 99 characters were parsimony-informative and 211 characters were parsimonyuninformative. The maximum parsimony analysis resulted in one tree (TL = 412, CI = 0.871, RI = 0.817, RC = 0.712, HI = 0.129) as shown in Supporting Figure 1. The tree was rooted with C. dematium CBS 125.25. The bootstrap support for the observed branching pattern is shown next to the branches. In the MP tree shown in Supporting Figure 1, Colletotrichum isolates (UASB- Cc-10, 20, 30, 40, 50, 60, 70, 73) clustered with the type strain of C. truncatum (CBS 151.35) within the C. truncatum clade. In order to confirm the affinity of MTCC and ITCC isolates to C. truncatum clade, MP analysis was performed for the ITS dataset. The dataset included 56 sequences and a total of 524 positions. Two characters from the ambiguous regions were excluded from the analysis. Out of the remaining 522 characters, 478 characters were constant, 6 characters were parsimonyinformative and 38 characters were parsimony-uninformative. The maximum parsimony analysis resulted in one tree (TL = 47, CI = 0.979, RI = 0.944, RC = 0.924, HI = 0.021) as shown in Supporting Figure 2. The tree was rooted with C. dematium CBS 125.25 as outgroup. The bootstrap support for the observed branching pattern is shown next to the branches. In the MP tree shown in Supporting Figure 2, all the ITCC and MTCC isolates clustered with the type strains of C. truncatum CBS 151.35 within C. truncatum clade