Figure S1 - Springer Static Content Server
advertisement
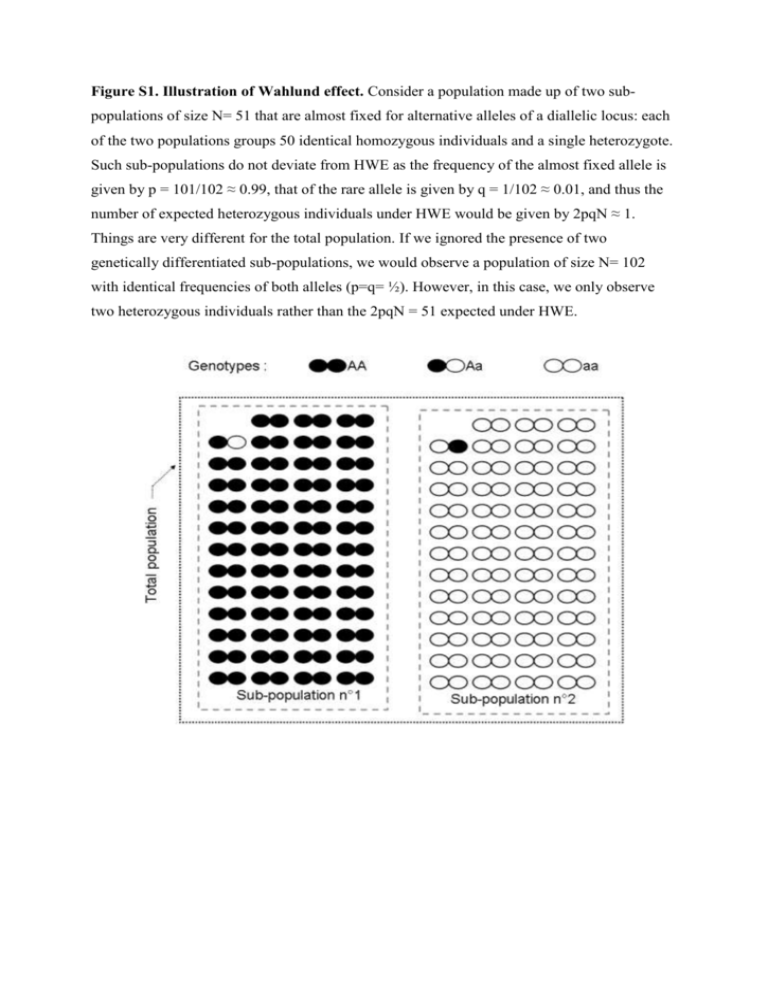
Figure S1. Illustration of Wahlund effect. Consider a population made up of two subpopulations of size N= 51 that are almost fixed for alternative alleles of a diallelic locus: each of the two populations groups 50 identical homozygous individuals and a single heterozygote. Such sub-populations do not deviate from HWE as the frequency of the almost fixed allele is given by p = 101/102 ≈ 0.99, that of the rare allele is given by q = 1/102 ≈ 0.01, and thus the number of expected heterozygous individuals under HWE would be given by 2pqN ≈ 1. Things are very different for the total population. If we ignored the presence of two genetically differentiated sub-populations, we would observe a population of size N= 102 with identical frequencies of both alleles (p=q= ½). However, in this case, we only observe two heterozygous individuals rather than the 2pqN = 51 expected under HWE. Figure S2. Comparisons of locus-specific estimates of F-statistics. For each F-statistic estimate, population genetics software may compute confidence intervals. Here, we present a putative analysis based on the polymorphism of nine independent genetic markers (i.e., in linkage equilibrium). The means and standard errors of the per locus estimates of the parameters FST (on left, estimator θ) and FIS (on right, estimator f) are displayed for the same set of populations for the nine independent loci. Seven loci provide congruent estimates for both parameters: they are thus likely to provide the information we are looking for, that is the population structure that results from the balance among genetic drift, migration, mutation and reproduction. Loci L8 and L9 provide different parameter estimates compared to the other seven and should thus be considered with caution; the estimates driven for these loci are likely to be biased by either the presence of null alleles and/or the action of selection pressures. 2 Supplementary file- Glossary: Bottleneck: Population bottleneck refers to drastic reduction in population sizes. This induces, at all loci, a decrease in genetic diversity and an even more pronounced drop in the allele numbers observed relative to a demographically stable population with the same census size. In other words the bottlenecked population will remain away from mutation/drift equilibrium for a number of generations following the reduction in census size. The more generations the bottleneck lasts, the stronger the demographic reduction, and the more intense and durable the genetic signature of such an event will be. Epistatic effects: This occurs whenever expression of a given gene A modifies expression of a gene B and vice versa. Fixation: An allele has reached fixation in a population when it remains the only allele present within a population. The immigration of different alleles into the population and mutation into a different allelic state are the only ways of re-creating local polymorphism in a fixed population. Hardy-Weinberg expectations: By definition, allelic and genotypic frequencies are expected to remain constant across generations within a population being at Hardy-Weinberg equilibrium (HWE). Complementary, under HWE, allelic frequencies are fully determined by the genotypic frequencies and vice versa. Consider one population of a diploid species and a molecular marker with two alleles A1 and A2 present at frequencies p and q = 1 - p, respectively. The frequency of A1A1 homozygous individuals will be given by the probability of sampling the A1 allele twice within the population, thus p². The frequency of A2A2 homozygous individuals will be determined by the probability of sampling the A2 allele twice, thus q². The frequency of A1A2 heterozygous individuals will be given by the probability of first sampling allele A1, then sampling the A2 allele, or of sampling them in the reverse order, thus pq + qp = 2pq= 1 - p² -q². If the population is not panmictic (everything else equal), the genotypic frequencies A1A1, A2A2 and A1A2 are given by [p²+pqFIS], [q²+pqFIS] and [2pq(1-FIS)], respectively. Genetic drift: A finite population size implies that, among all possible offspring that could be produced by a parental generation, only a sub-sample will occur and survive. This sampling effect, referred to as genetic drift, induces random fluctuations in allelic frequencies across generations until a point in time when, by chance (and in the absence of mutation or migration), one allele will be fixed in the population. As genetic drift is a random effect, it reduces the probability that nearby populations display the same distribution of allelic frequencies (i.e., it increases the genetic differentiation among populations). Its effect is 3 proportional to the effective size of the population; the smaller the population, the greater the effect of drift on allele frequencies. Linkage (dis)equilibrium: Linkage equilibrium refers to the independence between the polymorphisms observed at two loci. This implies that knowledge of an individual’s genotype at one locus provides no information on its genotype at the other locus. For the sake of simplicity, let us consider two bi-allelic loci A (with alleles A1 and A2) and B (with alleles B1 and B2) so that the genotypic frequencies recorded within a population are given, at locus A, by pA1A1, pA1A2 and pA2A2, and at locus B, by pB1B1, pB1B2, and pB2B2. These two loci are in linkage equilibrium whenever the frequencies of the nine bi-locus genotypes are determined by the product of the two mono-locus genotypic frequencies: pA1A1B1B1 = pA1A1 x pB1B1, pA1A2B1B1 = pA1A2 x pB1B1, pA2A2B1B1 = pA2A2 x pB1B1, pA1A1B1B2 = pA1A1 x pB1B2, pA1A2B1B2 = pA1A2 x pB1B2, pA2A2B1B2 = pA2A2 x pB1B2, pA1AB2B2 = pA1A1 x pB2B2, pA1A2B2B2 = pA1A2 x pB2B2, and pA2A2B2B2 = pA2A2 x pB2B2. The converse – linkage disequilibrium – refers to the case where there is some correlation between the polymorphism recorded at locus A and that recorded at locus B. There are several possible causes for linkage disequilibrium. Physical linkage among the considered loci is one reason: in this case, recombination among the two loci is not frequent enough to make the two mono-locus genotypes evolve independently from one another. Clonality and selfing generate linkage disequilibrium throughout the genome of the considered species. Selection may create linkage disequilibrium through epistatic effects, i.e., when the intensities of selection pressures acting on the polymorphism at a given locus A vary among the various genotypes observed at another locus B and vice versa. Finally, as an extension of the Wahlund effect when considering more than one locus, population structure creates linkage disequilibrium among loci, although not within subpopulations but at the level of the total (subdivided) population. Mutation/drift equilibrium: Consider an isolated population of constant finite size, and a neutral locus at which panmixia is realized so that it tends to be at HWE. At this locus along time, mutation will regularly introduce new alleles, whereas genetic drift will regularly drive some alleles to extinction. The combined action of mutation and genetic drift will lead the population to a dynamical equilibrium where the number of distinct alleles (hence the genetic diversity) remains constant. Panmixia: Zygote production follows random meeting of gametes produced in a population; theoretically achieved only for species where individuals are self-compatible hermaphrodites. Wahlund effect: Sampling errors so that individuals from distinct sub-populations are 4 merged within samples are described as Wahlund effects. Such effects result in distorting the relationships between the allelic and genotypic frequencies. An illustration is given in the supplementary material (Figure S1). 5