Linkagedocument
advertisement

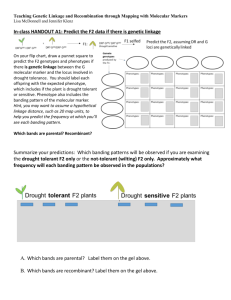
As of 02.14.03 we do not have a Pascal compiler for Linux, therefore there is no functional Linkage software for our Linux OS machines. One can install it in own working PC, by having in advance the PC compiler. (A.K.) This is an interesting note from http://bimas.dcrt.nih.gov/linkage/ltools.html#linkage LINKAGE Version 5.1, dated January 19, 1994: NOTE: It is strongly recommended that the use of the LINKAGE program be discontinued and FASTLINK (and/or VITESSE) be used in it's place. The functionality of FASTLINK is designed to supercede LINKAGE, and FASTLINK's performance benefits are very great. And for those problems which VITESSE can accomodate, the additional performance has the potential for being still greater. LINKAGE is an integrated system of programs designed to perform linkage analysis and genetic risk calculation for an arbitrary number of loci. These programs are subdivided as "Linkage Analysis" programs and "Linkage Support" programs. LINKAGE's Linkage Analysis programs Linkage Analysis programs consist of a core of programs, which perform such tasks as maximum likelihood estimation of recombination rates, calculation of lod score tables, and analysis of genetic risk. MLINK: This program performs n-point linkage and risk calculations in general pedigrees. This program can be used to construct lod score tables. LODSCORE: This program performs iterative maximum likelihood estimation of recombination fractions from two-locus data in general pedigrees. ILINK: This program performs iterative maximum likelihood estimations in general pedigrees. LINKMAP: This program calculates location scores in general pedigrees. UNKNOWN: This program infers possible genotypes for unknown individuals. Prepares input files for ILINK, LINKMAP, MLINK and LODSCORE programs. CILINK: This program performs iterative maximum likelihood estimations in nuclear pedigrees (nuclear families with the possible addition of grandparents). CMAP: This program calculates likelihoods for genetic location in nuclear pedigrees. CINFER: This program infers genotypes for unknown individuals whenever possible. Works on nuclear pedigrees only. Prepares the input file for the CFACTOR program. CFACTOR: This program applies factorization and transformation rules to genotypes in nuclear pedigrees prior to analysis with the CMAP and CILINK programs. CFACTOR works on nuclear pedigrees only. LINKAGE's Linkage Support Programs Linkage Support programs form an interactive "shell" or "front-end" around the core programs. In addition, they also provide the user with linkage file preparation, data checking facilities and report generation from the linkage results. LCP: Linkage Control Program (LCP) is an interactive menu program for building command files to perform linkage analysis. LRP: Linkage Report Program (LRP) is an interactive menu program for displaying and formatting the stream output from the linkage analysis programs. MAKEPED: converts pedigree files of the form: pedigree number, ID number, father ID, mother ID to a form that includes offspring and sibpointers. PREPLINK: interactively defines locus descriptions; stores descriptions in a parameter file for input into computational programs. LINKAGE references: G. M. Lathrop, J.-M. Lalouel, C. Julier, and J. Ott, Strategies for Multilocus Analysis in Humans, PNAS 81(1984), pp. 3443-3446. G. M. Lathrop and J.-M. Lalouel, Easy Calculations of LOD Scores and Genetic Risks on Small Computers, American Journal of Human Genetics, 36(1984), pp. 460-465. G. M. Lathrop, J.-M. Lalouel, and R. L. White, Construction of Human Genetic Linkage Maps: Likelihood Calculations for Multilocus Analysis, Genetic Epidemiology 3(1986), pp. 39-52. For more information, look at genetic linkage analysis software at Rockefeller University.