Directorate Learning Development

Department for Learning Development
’Students as Researchers Scheme’
**Please note this scheme is only available to undergraduate students**
Name
Section/Subject
School/Department
Dr Caroline Orr and Dr T. Komang-Ralebitso Senior
Biological Sciences – Molecular Microbial Ecology
Life Sciences Subject Group, School of Science and
Engineering c.orr@tees.ac.uk
; k.ralebitso-senior@tees.ac.uk
Contact email
Title of research project/activity Response of nitrogen-cycling bacteria to biochar addition
Pedagogic or Discipline Specific Discipline Specific – This project would link to the Biological
Sciences and Environmental Science undergraduate programmes
Summary of project research activity
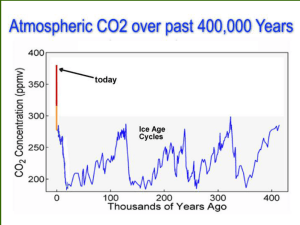
Within soil, bacteria play an essential role in the cycling of important nutrients such as organic carbon and nitrogen that contribute to plant growth and a range of associated ecosystem services. Perturbations of the soil have been shown to exert significant effects on the abundance and distribution of many groups of soil bacteria. Recent advances in molecular microbial ecology offer the opportunity to study key functional groups in the soil at a meaningful resolution.
Application of biochar to soil has been previously shown to promote plant growth and clean up contaminated sites through increases in total carbon, nutrient retention/availability, soil moisture holding capacity/permeability, organic matter and pH, and promoted microbial activity. However, few definitive studies have reported how the functional bacterial community is affected by biochar addition. As nitrogen is often the key-limiting factor to crop growth, understanding the effect of biochar addition on nitrogen fixation will be key to understanding how biochar may alter the dynamics of this essential element within soil.
Since, less than 5% of all microorganisms can be cultured in the laboratory; the research team are aiming to use a combined application of findings from published literature, including work already carried out within the University, and novel investigations that entail quantitative polymerase chain reaction (qPCR), denaturing gradient gel electrophoresis
(DGGE) and sequencing.
Previously, our research group established pot experiments comparing soil with and without biochar in the presence and absence of clover. Soil samples have been collected and
DGGE analysis has started to unpick how biochar affects the diversity of bacteria within the samples. However, we require the input of an enthusiastic and hard-working student to perform further DNA extractions, help develop a high-impact qPCR protocol and assess the abundance of the functional nitrogen cycling genes in response to biochar addition.
Have you obtained ethical clearance for this project?
Summary of student tasks
Research output(s) for student
Are there specific criteria students need to address when applying for the project?
Anticipated timeline for project
Number of student hours required (up to 65)
Are there any additional costs associated with the student researcher i.e. travel? Please
This is a low risk project in terms of ethical clearance hence it has previously been cleared by the SSE School Research
Ethics Committee. However, some small modifications have been made to the protocol. Therefore, the project will be resubmitted for consideration at the next ethics board
(04/02/2015).
The appointed student will:
1. Extract DNA from the soil samples already obtained.
2. Identify genes involved in nitrogen cycling using conventional PCR
3. Develop a working protocol for quantifying nitrogen cycling genes using qPCR
4. Apply statistical analysis to the abundance data to deduce the effect of biochar on the activity of the nitrogen cycling community.
It is envisaged that the researcher will be a Level 5 or Level 6
BSc (Hons) Biological Sciences/Environmental Science student. Therefore, s/he will be familiar with the DNA extraction, PCR and DGGE methodologies as their theoretical and practical principles are part of the curriculum in several modules including: Cell Biology and Microbiology;
Genetics and Molecular Biology; Molecular Ecology;
Ecological Sustainability. This research programme will afford opportunities to:
1. Hone technical and laboratory skills on molecular analysis
2. Improve competency in PCR
3. Introduce students to qPCR, allowing them to add to their molecular toolkit
4. Become co-author to a paper and book chapter currently in draft
5. Develop overall research proficiency as a young scientist
6. Gain insight into the postgraduate research environment
Students should show, and be able to provide evidence for:
1. An interest in molecular microbial ecology
2. A critical understanding of PCR
3. A desire and motivation to learn a new skill
4. An ability to manage themselves and their time
As indicated, the protocols for DNA extraction, PCR, agarose gel electrophoresis and DGGE will be familiar to the student.
Therefore, the parts of the project that require these techniques/skills will take place prior to the exam period.
During the exam period the student will not be expected to work on the project. The qPCR work will be new to the student and will take place following the exam period.
A Gantt Chart has been included to demonstrate the work plan.
65.
The project will require consumables for use in the laboratory. Funding has already been secured via the
University Research Fund for this.
provide details of how this will be funded