Unit 4 Review: Molecular Genetics
advertisement

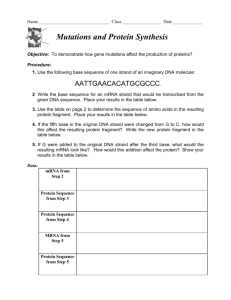
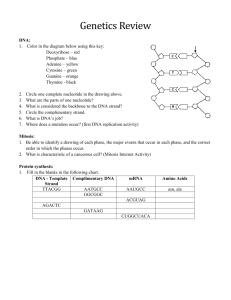
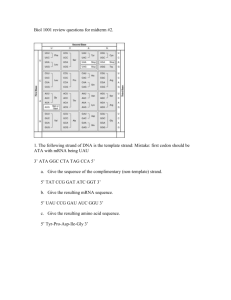
Unit 4 Review: Molecular Genetics 1) Fill in the following chart, which highlights the scientist who helped to establish DNA as the genetic material. Investigator(s) Experimental Organisms/Design and Conclusions S vs R strain of bacteria; something in heat-killed dangerous bacteria transformed Griffith living not dangerous bacteria into killer bacteria. Implied not a protein because heat would have denatured the protein. Avery Hershey and Chase Tested one component of Griffith’s experiment at a time; only DNA transformed the non-dangerous bacteria into killer bacteria Radioactively labeled P in 1 batch of virus and S in a 2 nd batch of virus; let each group infect a separate colony of bacteria. The only bacteria that had radiation inside where the group infected by radioactively labeled Phosphorus (P in DNA) Chargaff -Each species has a unique % of A, T, C, & G -% of C = % of G and % of T = % of A Franklin Made the diffraction picture of crystallized DNA that Watson & Crick used to help them identify the structure of DNA as a double helix 2) Summarize the evidence and techniques Watson and Crick used to deduce the double helix structure of DNA. -model building (wire & sticks) -used Rosalind Franklin’s X-Ray diffraction picture which showed the width of the helix (2nm), distance between base pairs (0.34nm), & the length of a full twist (3.4nm) 3) Label the following structure of DNA using the list below: nucleotide cytosine guanine adenine thymine purine base pyrimidine base 5’ end of chain sugar-phosphate backbone 3’ end of chain hydrogen bonds deoxyribose phosphate group a) sugar-phosphate backbone b) 3’ end c) pyrimidine d) purine e) H- bond f) cytosine g) guanine h) adenine i) thymine j) 5’ end K) nucleotide l) deoxyribose m) phosphate group 4) In the following diagram showing the replication of DNA, label the following items: leading strand, lagging strand, Okazaki fragment, DNA polymerase, DNA ligase, Helicase, Primase, single-stranded binding proteins, RNA primer, replication fork, and 5’ and 3’ ends of parental DNA. a) helicase b) single-stranded binding protein c) DNA polymerase III d) leading strand e) lagging strand f) DNA ligase g) DNA polymerase I h) Okazaki fragment i) primer j) replication fork K) primase l) 3’ end m) 5’ end 5) Determine the amino acid sequence for a polypeptide coded for by the coded for by the following DNA antisense strand (HINT: first determine the mRNA sequence that will be transcribed). DNA 3’- T A C G G A C T G A A A T T C A C T- 5’ mRNA 5’- A U G C C U G A C U U U A A G U G A- 3’ a.a. seq Met- Pro- Asp- Phe- Lys- STOP 6) What determines if a ribosome remains free in the cytosol? If the beginning of the amino acid sequence is recognized by a SRP (signal recognition particle), the SRP binds to the elongating amino acid chain and drags the entire unit (elongating amino acid chain & ribosome) to the rough ER and binds the ribosome to the rough ER. If the initial amino acid sequence isn’t recognized by the SRP, it stays “free” in the cytosol. 7) Using the chart in your textbook (p. 314), fill in the following table: DNA triplet GGG CTT TTT or TTC AGC GAA, GAC, AAT, GAG, GAT, AAC ATA AAA GTT mRNA codon CCC GAA AAA or AAG UCG CUU, CUG, UUA, CUC, CUA, UUG UAU UUU CAA tRNA Anticodon GGG CUU UUU or UUC AGC GAA, GAC, AAU, GAG, GAU, AAC AUA AAA GUU Amino Acid Pro Glu Lysine Ser Leucine Tyr Phe Gln 8) Define the following and explain what type of point mutation could cause each of these mutations: a) silent mutation: base-pair substitution with no effect (ex: the amino acid is the same due to redundancy of the code or the amino acid is really similar in chemical properties) b) missense mutation: substitute base-pair with some effects (new amino acid but some differences in folding; ex: hemoglobin & Sickle Cell makes sense, just not the “right” sense c) nonsense mutation: a point mutation results in a STOP codon; translation ends prematurely d) frameshift mutation: insertion or deletion that changes the reading frame (groups of 3 mRNA nucleotides); as a result, all amino acids are wrong after the mutation 9) Eukaryotic cells modify mRNA after transcription. Describe how the pre-mRNA is modified with respect to: a) the 5’ ends and 3’ ends 5’ end: modified G cap 3’ end: poly A-tail b) RNA splicing of interior sections (introns vs. exons) Spliceosome (snRNA & proteins): splice/cut out Introns & join Exons into one continuous coding sequence 10) Sketch and describe the anatomy of a ribosome. Include in your labeled sketch and/or your description the following: large ribosomal unit (50s), small ribosomal subunit (30s), E site, P site, A site, mRNA binding site, and growing polypeptide.