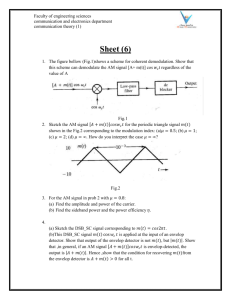
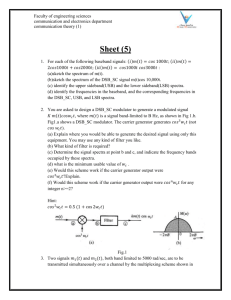
Supplemental Figure Legend
advertisement

Legend for Supplementary Figure 1 We examined DSBs in the region around the URA3-ARG4 interval in the following rad50S strains. MJL2960 carries the natMX, hphMX, ura3-EcPal+104 and arg4EcPal+9 markers in the same configuration as strains (MJL2936 and MJL2957) that were used for the genetic analysis. Strain MJL2961, which was analyzed in Figure 3 of the main text, carries only the URA3-ARG4 interval at HIS4 without palindromes and without the flanking natMX or hphMX markers. In part A of this figure, DNA was isolated from MJL2960 at 0 and 7 hours after transfer to sporulation media. The DNA was then digested with XhoI (X), and the subsequent Southern blot was probed with ARG4 sequences. The diagrams next to the Southern blots in parts A and B use the same symbols as Figure 2 of the manuscript but are not drawn to scale. In this particular gel of strain MJL2960, 27.4% of insert-containing chromosomes have a DSB at the hotspot in the URA3-ARG4 interval. In MJL2961 we observed 20% of insert containing chromosomes with a DSB at the same hotspot (Figure 3A and 3C). Additional DSBs were observed in MJL2960 (part A) that were absent in MJL2961 (not shown). These DSBs map to each side of the natMX marker and are present in 2.1% and 0.3% of insert containing chromosomes. Part B shows a trace of intensities from the 0 (black line) and 7 (red line) hour time points. In part C of this figure DNA from MJL2960 and MJL2961 was digested with PpuMI (P) and the subsequent Southern blot was probed with ARG4 sequences. No DSBs are detected in the region flanking the URA3-ARG4 interval on the hphMX side Jessop et al. Online legends -1 (21.6 kb). Part D shows the trace of intensities from the 0 (black line) and 5 (red line) hour time points from MJL2960. From these data we conclude: 1. the presence of the palindromes and flanking drug markers have little effect on DSBs in the URA3-ARG4 interval. 2. approximately 2.4% of insert containing chromosomes (2.1 + 0.3%) have a DSB at natMX and no other DSBs occur on the right side of the URA3-ARG4 interval in the 14 kb XhoI restriction fragment. Since the 14.0 kb and 12.7 kb parental bands were resolved on this gel, we believe that we would have detected DSB bands of 11.4 kb or less. 3. We estimated less than 0.7% of insert-containing chromosomes contain a DSB in about 13 kb to the left of the URA3-ARG4 DSB hotspot. This estimate comes from treating the spot near the MJL2960 5 hr lane as a DSB, and measuring its intensity. Other probings of the same blot indicate that a band of 13.1 kb was resolved from the 20.4/21.6 kb parental PpuMI bands. Therefore, we believe that we would have detected DSB bands of 13 kb or less. 4. Therefore, at least 90% [27.4/(27.4+2.4 + 0.7)] of DSBs in a region of 23 kb surrounding the URA3-ARG4 hotspot occur at the URA3-ARG4 DSB hotspot. Legend for online Appendix Jessop et al. Online legends -2 Patterns of marker segregation in MJL2902, MJL2870, MJL2936 and MJL2957. Parental genotypes, are given at the top of each page and below. MJL2902: --HIS4 URA3 ARG4 leu2; --his4 ura3 arg4 LEU2. MJL2870: --his4 URA3 arg4 LEU2; --HIS4 ura3 ARG4 leu2. MJL2957, MJL2936: --natS URA3 ARG4 HYGR; --NATR ura3 arg4 hygS. Spores showing PMS are shown as circles that are half black and half open. For each tetrad type, the following information is given. The number to the left of each tetrad indicates the number of tetrads with the pattern of segregation shown. On the right, the type of gene conversion is noted. If the conversion is CO-associated, the interval where the exchange point was mapped is noted by a Roman numeral (see text). Incidental COs are noted by an italicized Roman numeral. In ectopic crosses (MJL2902 and MJL2870), allelic CO between HIS4 and LEU2 can generate the same marker configuration as some ectopic COs. These allelic COs (denoted “allelic”) were identified as described in Materials and Methods. For MJL2902, MJL2957 and MJL2936, single spores that showed co-PMS were scored as having either cis or trans co-sectoring (indicated in red boldface type). Spores with cis co-sectoring contain one parental genotype for both markers in one colony sector, and the other parental genotype in the other colony sector. These are indicated with half-filled circles in the same orientation. Spores with trans co-sectoring contain non-parental genotypes in both sectors, and are indicated with half-filled circles in opposite orientations. Co-sectoring patterns were not recorded for MJL2870; thus, all tetrads with co-PMS in a single spore are denoted “cis/trans” in red boldface type. CoJessop et al. Online legends -3 PMS tetrads with sectoring patterns consistent with the DSBR model are indicated in red type as “DSBR CO” or “DSBR NCO”. Co-PMS tetrads with sectoring patterns that must have been produced by multiple events are denoted “multiple” in red boldface type. Tetrads showing co-conversion of the palindromic markers have been divided into two categories, “simple” and “complex”. Simple co-conversions appear to have resulted from a single event. Complex co-conversions are likely to have resulted from multiple events. Complex co-conversions are further divided into two categories, ”simple + other” and “multiple”. “Simple + other” tetrads contain more than two recombinant chromatids, but at least one pair of recombinant chromatids could have been produced by a single event. Tetrads in the multiple sub-category contain segregation patterns that cannot be explained by a single DSBR event (see text). We note that there is often more than one possible interpretation of the pattern of marker segregation in these tetrads; our goal was to choose the interpretation with the fewest possible events. In cases where a marker segregation pattern could be explained by either a two-strand double crossover or two gene conversion events, we chose the former. Choosing the latter explanation slightly decreases the number of crossovers and slightly increases the number of conversion events recorded, but does not materially alter any conclusions (data and analysis not shown). Jessop et al. Online legends -4