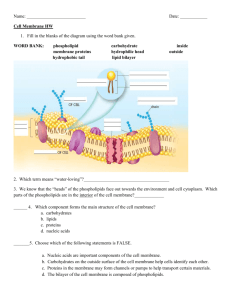
in word format - Department of Biological Sciences
advertisement

AOHUPO Membrane Proteomics Workshop 8 December 2006 Venue: University of Singapore, LT 32, Science Drive 4 This Workshop and the AOHUPO Membrane Proteomics Initiative are proudly sponsored by GE Healthcare 9:00- 10:00 Session 1 9.00-9.20 Bill Jordan (Centre for Biodiscovery, Victoria University of Wellington, Wellington) “The AOHUPO Membrane Proteomics Initiative” 9:20-9.40 Fuchu He (Beijing Proteome Research Center, Beijing Institute of Radiation Medicine, and Institutes of Biomedical Sciences, Fudan University, Shanghai) “Proteomic analysis of endoplasmic reticulum in C57BL/6J mouse liver” 9:40-10:00 Young-Ki Paik (Yonsei Proteome Research Center, Yonsei University, Seoul) “Proteomic analysis of rat liver microsomal proteins by a liquid-based twodimensional chromatography combined with nano-LC-ESI-MS/MS” 10:00-10.30 Morning tea 10:30-11.30 Session 2 10:30-10:50 Paul Michael (Technology Manager, Molecular Biology GE Healthcare, UK) “Multi-parameter cellular analysis” 10.50-11.10 Rudolf Grimm (Agilent Technologies Inc., Santa Clara, California) “A novel strategy to enhance membrane protein identification by reversed-phase protein prefractionation and HPLC-Chip/MS” 11:10-11:30 Lifeng Peng (Centre for Biodiscovery, Victoria University of Wellington, Wellington) “Proteomic analysis of a microsomal subcellular fraction from lactating bovine mammary gland” 11.30-12:10 Session 3 11:30-11:50 Alice Len (Australian Proteomics Analysis Facility, Sydney) “Membrane protein shotgun peptide IEF” 11.50-12:10 Robert Goode (Joint ProteomicS Laboratory, Ludwig Institute for Cancer Research / Walter & Eliza Hall Institute of Medical Research and Department of Biochemistry and Molecular Biology, University of Melbourne, Mellbourne) “The cationic colloidal silica strategy for purification of adherent, integral, plasma membrane proteomes from epithelial cells” 12:10 Discussion “Membrane proteomics and the AOHUPO Membrane Proteomics Initiative” 1:00 End of Workshop 1:00 Lunch ABSTRACTS THE AOHUPO Membrane Proteomics Initiative Bill Jordan, Lifeng Peng, Magalie Boucher, Graeme Lindsay, Pisana Rawson, Daniel Klotz, Florian Dreyer Centre for Biodiscovery and School of Biological Sciences, Victoria University of Wellington, PO Box 600, Wellington 6140, New Zealand. AOHUPO has chosen membrane proteomics as a research theme. This Membrane Proteomics Initiative (MPI) is relevant to the larger goals of HUPO in characterising the human proteome and to the biologically more diverse interests of the countries in the AOHUPO region. There is therefore a human focus but also relevance to microbial, plant and animal proteomics. This initiative was selected because of the importance and challenge of membrane proteomics. Membrane proteins are calculated to represent a large fraction of genomes, are essential in processes including signal reception and transduction, are implicated in disease and are important targets for industry. Further information is available at www.aohupo.org Phase I of the MPI is technology development. A membrane standard has been prepared for distribution to the participating laboratories. A range of proteomics methods will be tested and compared with the goal of defining optimum analytical methods for the various kinds of membrane proteins including trans-membrane proteins with multiple membrane spanning domains. The MPI standard has been prepared from the large lobe of livers from male 10-11 week old C57BL/6J mice. Preparation and distribution of this standard has been generously supported by GE Healthcare. Proteomic analysis of endoplasmic reticulum in C57BL/6J mouse liver Yanping Song1, Ying Jiang1, Fuchu He 1,2 1. Beijing Proteome Research Center, Beijing Institute of Radiation Medicine, 33 Life Science Road, Beijing 102206, China. 2. Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China Endoplasmic reticulum (ER) takes part in the protein synthesis, glycogenolysis and biosynthesis of lipid and cholesterol. It also has a close relationship with membranebound and secretory proteins. For liver tissue, ER plays a great role in the biotransformation of hydrophobic endogenous metabolites and xenobiotics. The disorder of ER function can cause cell dysfunction and diseases. To gain a better understanding of the critical role of ER function in liver, we present the ER proteome study on C57BL/6J mouse liver. In contrast to other proteome studies we focused on the multiple fractionation strategy to make full use of sample and to compare different subcellular fractions from single homogenate as well. The purity evaluation method was also applied to obtain highly purified organelle. Subsequently, ER proteins were separated by SDSPAGE prior to nano-LC-ESI-MS/MS analysis with gas phase fractionation by mass range selection. 2018 and 1735 proteins were identified separately in rough and smooth endoplasmic (rER, sER), in which 1232 and 1177 proteins matched by over two peptides. Proteomic analysis of rat liver microsomal proteins by a liquid-based twodimensional chromatography combined with nano-LC-ESI-MS/MS Hyoung-Joo Lee; Min-Seok Kown; Eun-Young Lee; Young-Ki Paik Yonsei Proteome Research Center, Yonsei University, Seoul, Korea A liquid-based two-dimensional separation system (2D-LC) method combined nano-LCESI-MS/MS was developed for analysis of hydrophobic rat liver microsomal proteins. The membrane proteins were separated on chromatofocusing as neutral pH range (pH 8.5-4.0) and basic range(10.5-7.4) for the first dimension followed by non porous reversed-phase HPLC for the second dimension. The detergent 2 % Trion X-100 was used for efficient solubilization of membrane proteins and removal of soluble protein. Nano-LC-ESI-MS/MS was used for large-scale identification of fractionated proteins. Among 140 proteins identified by nano-LC-MS/MS, about 48% proteins were hydrophobic proteins with more than one transmembrane domain (TMD) up to proteins with 15 TMDs. Approximately 28 % of identified proteins was located on membrane region. With respect to function, 28% proteins were identified to be involved in metabolism, 14% protein were related to cell signal and regulation and 14% proteins involved in transportation. The 2-D virtual image of pI vs. hydrophobicity could be used for differential display analysis. Thus, our 2D-LC system can provide large-scale analysis of hydrophobic rat liver microsomal proteins not only for the analytical separation but for the preparative separation with high resolution as compared to traditional gel methods. [supported by MOHW grant 03-PJ10-PG6-GP01-0002 (to YKP)] Multi-parameter Cellular Analysis Paul Michael Technology Manager, Molecular Biology, GE Healthcare, UK High-content analysis offers a range of approaches for high-throughput cellular analysis providing an in-depth view of the effects of genes and drugs on biological processes. A combination of automated imaging, sophisticated software and complementary fluorescent biology are required to enable high-content analysis. This presentation will cover validated biological assays and algorithms developed on robust imaging platforms. A novel strategy to enhance membrane protein identification by reversed-phase protein pre-fractionation and HPLC-Chip/MS R. Grimm Agilent Technologies Inc., 5301 Stevens Creek Blvd, Santa Clara, CA95052, USA It is predicted that more than one third of the open reading frames in the human genome are encoded with membrane proteins. Membrane proteins are involved in many crucial biological processes and scores of examples have shown a direct link to human diseases, thus there is an increasing interest to study membrane function and apply the knowledge to drug discovery. However, analysis of membrane proteins represents a major challenge in proteomics due to their amphipathic nature, and despite their biological importance they remain an underrepresented subset of studied proteins within proteomics. We have developed a strategy for enhancing membrane protein identifications by two dimensional pre-fractionation and microfluidic nano-chip-LC/MS analysis. Specifically, we have used a novel high-recovery macroporous reversed-phase (RP) C18 column to pre-fractionate intact HeLa membrane proteins under high temperature conditions. Collected RP fractions were further resolved by SDS-PAGE, in-gel digested and the resulting tryptic peptides analyzed by HPLC-Chip/MS. Alternatively, RP fractions were in-solution digested and tryptic digests analyzed via 2D HPLC-Chip/MS. To date, we have identified more than 900 proteins with more than half of the identifications predicted or verified as integral membrane proteins. This robust and highly reproducible workflow has demonstrated utility for increasing membrane protein identifications from a highly complex membrane protein sample. Optimized conditions for the RP separation permitted total protein recoveries greater than 95%, enhanced peak resolution and showed excellent reproducibility. Proteomic analysis of a microsomal subcellular fraction from lactating bovine mammary tissue Lifeng Peng, Pisana Rawson, Danyl McLauchlan and T. William (Bill) Jordan School of Biological Sciences, Victoria University of Wellington, Wellington, New Zealand. A proteomic investigation of a microsomal subcellular fraction from lactating bovine mammary tissues was carried out using 2-DE with MALDI TOF MS and 1-DE with RPLC ESI MS MS. 2-DE allowed 210 protein identifications, of which 45 (21.5%) were microsomal proteins and 39 (18.6%) membrane proteins including 17 transmembrane proteins. 1-DE revealed 723 proteins, of which 211 (29.3%) were microsomal proteins and 204 (28.3%) were membrane proteins including 167 transmembrane proteins. From the point of view of biological functions, 50 (23.2%) of the microsomal proteins are involved in protein biosynthesis, 64 (30.4%) in protein folding, trafficking, assembly, sorting and secretion and 22 (10.3%) were related to lipid metabolism, transport and secretion, which is consistent with the prediction of the biological functions of microsomes. Comparison of 2-DE and 1-DE results shows that the performance of 1-DE in combination with RPLC ESI MS/MS was much greater than that of 2-DE with regard to membrane proteins. The data reported here presented a comprehensive proteome survey of the enriched microsomal fraction from the lactating bovine mammary tissues, which provides knowledge for understanding molecular mechanisms in mammary glands. Exploiting the membrane subproteome. A shotgun peptide immobilised pH gradient – isoelectric focusing approach* Joel Chick1, Mark Molloy1,2, Mark S. Baker1,2, Alice Len1,2 1 Dept. Chemistry & Biomolecular Sciences, Macquarie University, NSW, 2109, Australia. 2 Australian Proteome Analysis Facility Ltd, Macquarie University, NSW, 2109, Australia. Membrane proteins play an important role in pathogenesis and are one of the most exploited targets for therapeutic intervention. Proteomics is contributing to improving the understanding of many disease processes at a molecular level. Despite this, traditional proteomic methods used to analyse and identify membrane proteins remain a significant bottleneck where there has been slow progress. Overcoming physicochemical limitations dictated to by the hydrophobic nature of the vast majority of membrane proteins is a key factor to enhancing the analysis of this repertoire of proteins. We address this inherent limitation by employing a shotgun proteomic approach. An established form of this technique involves tryptically digesting a complex mixture of proteins and separating the generated peptides over single or multidimensional liquid chromatography mass spectrometry. Here, we present preliminary data obtained from analysing rat liver membrane proteins utilising a peptide immobilised pH gradient - isoelectric focusing coupled with this shotgun proteomic approach. Preliminary analysis of a total of 24 fractions of peptides focused over a pH range of 3-10 yielded an identification of 2957 non redundant peptides, corresponding to a total of 761 unique proteins with a false positive rate of 1%. 51 of these proteins have a GRAVY score of > +0.3, validating the suitability of the approach for hydrophobic proteins that are often found to be refractory to analysis by 2DE gels. Some examples of the rat liver membrane proteins identified using this approach include putative transferrin receptor protein 2 (TfR2), solute carrier family proteins and many cytochrome P450 genetic variants, to name but a few. This study has illustrated that the majority of rat liver tryptic peptides focused within the pH range of 3.3-5.2. Further in-depth analysis of the peptides within this narrow IPG strip pH range will facilitate better peptide resolution and subsequently enable further coverage of the membrane subproteome. * The authors would like to acknowledge GE Healthcare for their support as a industry partner for the ARC linkage grant LP 0561182 that supports this project. The cationic colloidal silica strategy for purification of adherent, integral, plasma membrane proteomes from epithelial cells Robert Goode1,2, Eugene Kapp1, Robert Moritz1 and Richard Simpson1,2 1 Joint ProteomicS Laboratory, Ludwig Institute for Cancer Research / Walter & Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia. 2 Department of Biochemistry and Molecular Biology, University of Melbourne, Parkville, Victoria, Australia. The plasma membrane is a desirable source for discovery of both biomarkers and drug targets. However, purification of the plasma membrane specifically is often troublesome due to the relative abundance of other cellular membranes and variability in membrane density between cell lines. Additionally, classical membrane preparations are often heavily contaminated with abundant basic proteins, such as histones and ribosomal proteins (Simpson et al., 2000). Here we present a method for the purification of the adherent plasma membrane from epithelial cell lines using cationic colloidal silica apical membrane subtraction (Goode and Simpson, in press) and demonstrate its specificity for integral plasma membrane proteins through detailed proteomic analysis. Of over 200 manually validated identifications, over 50% possess between 1 and 12 transmembrane domains, including over 30 CD antigens and several markers of the basolateral plasma membrane. Goode, R.J.A. and Simpson, R.J. (in press). "Purification of basolateral integral membrane proteins by cationic colloidal silica-based apical membrane subtraction" in Proteomic analysis of membrane proteins: Methods and protocols in Methods in Molecular Medicine series. Simpson, R. J., et al. (2000). "Proteomic analysis of the human colon carcinoma cell line (LIM 1215): Development of a membrane protein database." Electrophoresis 21(9): 1707-1732.