MeV Analysis of Gene Expression Data - 91-421-Spring-2009
advertisement

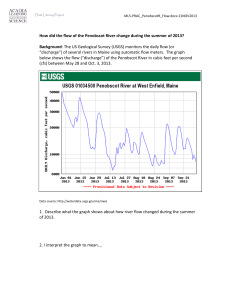
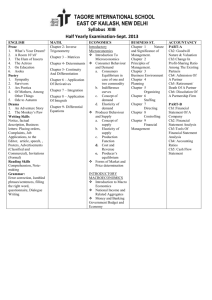
Week 2 Report Weka Filtering + ROC MeV Analysis of Gene Expression Data Spanish Inqusition (Yan Tran, Leon Kay, Chris Thomas) April 30, 2009 MeV Analysis of Gene Expression Data Data Preparation The gene expression data CSV file was transposed using a perl script and then converted to a tab-delaminated file so that it could be loaded into MeV. Upon loading a hierarchical clustering with both the gene tree and the sample tree options selected was performed. Observations The left side of the of the HCL tree data was dominated by samples that were Basal-like and the right side was dominated by samples that were either classified as Luminal-A or Luminal-B. Less than a quarter of the way down HCL tree data, there was a large area of genes that were mostly lowly expression on the left side and highly expressed on the right side. Figure 1: Initial MeV hierarchical clustering. The cluster of genes were selected and then opened in a separate MeV window. Another hierarchical cluster was then performed. It could then be clearly seen that the point where it goes from lowly-expressed to highly-expressed corresponds with the transition from Basal-like samples to Luminal-A/B samples. Figure 2: A cluster of genes that are lowly expressed on the left and highly expressed on the right. Internet searches were performed on the random genes listed in the cluster for more information on how they relate to breast cancer. The first one that yielded any information was GATA3. A paper that was found correlated directly with what was shown in MeV: Basal-like tumors have the lowest levels of GATA3 expression, while luminal-type tumors have the highest. The paper mentioned that basal-like tumors have the worst prognosis while luminal tumors have a better prognosis [1]. Also, GATA3 also had an association with estrogen receptor alpha levels. The estrogen receptor alpha gene is often highly expressed in the early stages of breast cancer. [2] GATA3 was grouped with FLJ13710 in the gene tree. A quick search of the Internet validated the MeV analysis which showed that, like GATA3, FLJ13710 there is a significant difference in the level of expression for this gene between luminal and basallike samples [3]. It is mentioned in a paper that discusses genes that serve as prognostic signatures for breast cancer, but unlike GATA3, not much more information could be found on this gene. [4] Figure 3: FLJ13710 and GATA3 Conclusions MeV is a useful tool for identifying attributes that may be associated with specific classifications. Weka Filtering + ROC For Weka filtering we used CFS with BestFirst Search. This reduced the number of attributes from 1544 down to 125. CFS stands for Correlation-based Feature Selection. This was originally introduced in a paper by Mark Hall. The basic hypothesis of CFS is this: “A good feature subset is one that contains features highly correlated with (predictive of) the class, yet uncorrelated with (not predictive of) each other.” [5] CFS uses a formula to rank the correlation (strong to class, weaker to other features is better) of all the features. Then, a search algorithm is used to search and find the best subset out of the ranked features. Any search algorithm can be plugged into CFS – the author describes three - forward selection, backward elimination, and best first. They are all essentially greedy heuristic search algorithms. The greedy search approach reduces the complexity of generating the feature subset. The author describes BestFirst search in this manner: “Best first can start with either no features or all features. In the former, the search progresses forward through the search space adding single features; in the latter the search moves backward through the search space deleting single features. To prevent the best first search from exploring the entire feature subset search space, a stopping criterion is imposed. The search will terminate if five consecutive fully expanded subsets show no improvement over the current best subset.” [5] Figure 4 shows the results of the accuracy of the 5 learning algorithms before and after the CFS/BestFirst Search Filtering. Before* After** Error Rate Reduction J48 32.17 28.02 12.92 Bagging (J48) 18.26 16.38 10.30 Boosting (J48) 20.87 16.38 21.52 Random Forests 15.65 14.22 9.12 SMO (SVM) 15.22 14.22 6.53 * From Week1 - all 1544 Attributes ** After applying CFS/BestFit filtering, 125 attributes Figure 4 - Before and After Filtering - Accuracy results of 5 learning algorithms. ROC graphs “depict the tradeoff between hit rates and false alarm rates of classifiers" [2]. Area Under Curve, or AUC is an accurate numerical value that can be used to compare classifiers. Shown below, in Figure 5, is the ROC values of all 5 learning algorithms/classifications after doing the CFS filtering in Weka. J48 Bagging (J48) Boosting (J48) Random Forests SMO (SVM) Basal-like Claudinlow 0.8978 0.9851 0.9883 0.9939 0.9802 0.9515 0.9993 0.9975 0.9979 0.9977 HER2+/ER- 0.8137 0.9614 0.964 0.9476 0.9313 Luminal A 0.856 0.9558 0.9497 0.9735 0.9418 Luminal B 0.7842 0.93 0.9183 0.9336 0.9563 Normal Breast-like 0.7676 0.9731 0.922 0.955 0.9772 Figure 5 - ROC values for the 5 learning algorithms after Weka CFS filtering References 1. Wilson, Brian J., Giguère, Vincent. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Molecular Cancer 2008, 7:49. http://www.molecular-cancer.com/content/7/1/49 2. Hayashi, SI., et al. The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application. http://erc.endocrinologyjournals.org/cgi/content/abstract/10/2/193 3. Suppl. Table 2: List of probe sets significantly differentially expressed between luminal cell lines and basal cell lines. Probe sets are ordered according to decreasing DS (discriminating score). www.nature.com/onc/journal/v25/n15/extref/1209254x4.xls 4. Carrivick, L., et al. Identification of Prognostic Signatures in Breast Cancer Microarray Data using Bayesian Techniques. http://www.enm.bris.ac.uk/cig/pubs/2005/rs4.pdf 5. Mark Hall, “Correlation-based Feature Selection for Machine Learning”, http://www.cs.waikato.ac.nz/~mhall/thesis.pdf 6. Tom Fawcett, “An introduction to ROC analysis“, doi:10.1016/j.patrec.2005.10.010 – enter into http://dx.doi.org/