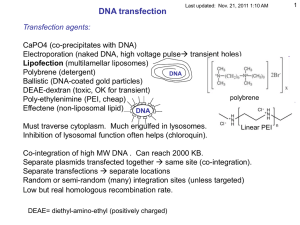
1 - UCSF Biochemistry & Biophysics
advertisement

You are a Drosophila geneticist studying yfg. In situ hybridization detects interesting expression patterns of yfg RNA during development. In each segment of the embryo, you observe yfg to be expressed in a diamond-shaped group of cells on the dorsal surface and in a spade-shaped group of cells on the ventral surface. In the larvae, you also find yfg to be expressed, but here it is in a club-shaped pattern on the wing disc. You decide to take a look at the genomic sequence near yfg, and find that there is 25 kb of sequence upstream of the gene that lacks any other annotated genes. You compare this to the genomic sequence of twelve other Drosophila species (http://flybase.org/blast/) and find stretches of 15 to 45 bp of perfect conservation of sequence. Furthermore, these islands of conserved sequences form three archipelagos that are each about 3 kb in size, separated by several kb of non-conserved sequence. You then probe for the pattern of expression in Drosophila pseudoobscura (one of the sequenced species that you looked at) and you find the embryonic diamonds and spades but the wing disc expression is now in the shape of a heart. Descriptions of sensory organ distribution in D. melanogaster and D. pseudoobscura mention a heart-like distribution in wings of D. pseudoobscura and a club-like distribution in D. melanogaster. You look at the sensory organ distribution in larvae and find a segmentally repeated diamond pattern on the dorsal side and a segmentally repeated spade pattern on the ventral side. A late larval lethal mutation mapping close to yfg was reported to have behavioral defects and you find that it lacks the ventral sensory organs. (1) You are intrigued! How would you test the hypothesis that each archipelago of conserved sequences acts autonomously to direct expression of yfg in each of its suit like shapes? (2) How would you test whether expression of yfg is sufficient to direct formation of sensory organs? (3) How would you determine whether the change in the distribution of yfg in the wings (i.e. club pattern in D. melanogaster and a heart pattern of D. pseudoobscura) was due to differences in cis-acting elements? Assume - that you have done screens to isolate mutants in the structural gene and in the various regulatory regions of yfg. - that all the mutations are lethal when homozygous except for ones that only alter the pattern of expression of yfg in the adult wing, and that these mutations alter the distribution of sensory organs in the wing. - that you have a deletion that covers yfg, both structural and regulatory regions. (4) How do you expect the mutations in the structural gene and in the regulatory regions to behave in complementation tests (a) with the deletion and (b) when tested against each other? (5) Given that mutation of the structural gene is lethal to the fly, how would you test whether yfg function is required for sensory organ formation? In your answers to these question please be fairly specific; if you use constructs, flies, crosses and analyses, indicate what they are but feel free to invent markers, promoters etc., within reason.