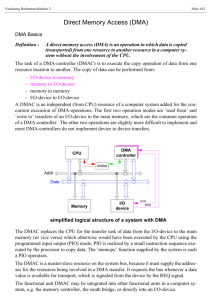
Colorimetric and Fluorimetric Fluoride Sensors Based on Novel
advertisement

Supporting Information Anion–π and CH···anion interactions in naphthalenediimide-based coordination complexes Atanu Mitra, Ronald J. Clark, Christian T. Hubley, and Sourav Saha* Department of Chemistry and Biochemistry and Integrative NanoScience Institute, Florida State University, 95 Chieftan Way, Tallahassee, Florida 32306, United States Email: saha@chem.fsu.edu Experimental Section MATERIALS DPNDI ligand was synthesized from 1,4,5,8-naphthalenedianhydride and 4-aminopyridine according to a literature procedure.S1 Zn(ClO4)2.6H2O, DMAc, and MeCN were purchased from commercial sources. Deuterated DMSO-d6 was purchased from Cambridge Isotope Laboratories was used for NMR studies. Synthesis All reactions were performed under N2 atmosphere using dry solvents unless otherwise specified. 1H was recorded in DMSO-d6 at 298 K in Bruker Avance 400 MHz spectrometers. Electrospray Ionization mass spectra (ESI-MS) were recorded on a JEOL AccuTOF JMS-T100LC ESI mass spectrometer. [Zn(DMAc)4–DPNDI–Zn(DMAc)4]4+ (1). A solution of DPNDI (55 mg, 0.13 mmol) in DMAc (14 mL) was added dropwise to a solution of Zn(ClO4)2.6H2O (48 mg, 0.13 mmol) in DMAc (6 mL) and the reaction mixture was allowed to stir at room temperature for 16 h. After a slow diffusion of Et2O vapour into the DMAc solution, off-white crystals precipitated out, which was used for the X-ray analysis. [Zn(II)–DPNDI]n coordination polymer (2). A solution of DPNDI (55 mg, 0.13 mmol) in 1:1 MeCN/DMAc (14 mL) was added dropwise into a solution of Zn(ClO4)2.6H2O (48 mg, 0.13 mmol) in 1:1 MeCN/DMAc (6 mL) and the reaction mixture was allowed to stir at room temperature for 16 h. After removing MeCN under reduced pressure to concentrate the reaction mixture, slow diffusion of Et2O into the DMAc solution yielded off-white crystals, which was used for crystallographic analysis. METHODS UV/Vis Spectroscopy UV/Vis spectra were recorded on a PerkinElmer Lambda-25 UV/Vis spectrophotometer. UV/Vis studies were conducted in DMSO, using the concentrations of 10 µM. Energy Minimized Structure of DPNDI/ClO4– Complex The energy minimization of DPNDI molecules and DPNDI/ClO4 complex were carried out applying the density functional theory (DFT) with the Becke three-parameter hybrid exchange functional in concurrence with the Lee-Yang-Parr gradient-corrected correlation function (B3LYP functional) with the 6-31+G** basis set [B3LYP/6-31+G**] using Gaussian 03 program.S2 Because of the electron withdrawing carbonyl groups and imide Ns, the imide rings are the most electron deficient part of NDI S1 molecules. Electrostatic potential surfaces (EPS) of NDIs (blue and red regions are electron deficient and electron rich region respectively in ESP) has been used to illustrate areas of low electron density that can interact with electron-rich anions. B3LYP/6-31+G** energy minimize structures of [DPNDI·ClO4–] shows that ClO4– is preferentially located on top of an imide ring carrying two electron withdrawing C=O bonds. One of the four O atoms of tetrahedral ClO4– anion preferentially interacts with DPNDI’s imide ring via anion–π interaction (d = ca. 3 Å) and another O atom display a CH···anion interaction (d = 2.3 Å) with the pyridyl ring, which creates a dihedral angle of ca. 80° with the core NDI plane. References: S1 S2 Guha, S.; Saha, S. J. Am. Chem. Soc., 2010, 132, 17674. Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Montgomery, Jr., J. A.; Vreven, T.; Kudin, K. N.; Burant, J. C.; Millam, J. M.; Iyengar, S. S.; Tomasi, J.; Barone, V.; Mennucci, B.; Cossi, M.; Scalmani, G.; Rega, N.; Petersson, G. A.; Nakatsuji, H.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Klene, M.; Li, X.; Knox, J. E.; Hratchian, H. P.; Cross, J. B.; Bakken, V.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R. E.; Yazyev, O.; Austin, A. J.; Cammi, R.; Pomelli, C.; Ochterski, J. W.; Ayala, P. Y.; Morokuma, K.; Voth, G. A.; Salvador, P.; Dannenberg, J. J.; Zakrzewski, V. G.; Dapprich, S.; Daniels, A. D.; Strain, M. C.; Farkas, O.; Malick, D. K.; Rabuck, A. D.; Raghavachari, K.; Foresman, J. B.; Ortiz, J. V.; Cui, Q.; Baboul, A. G.; Clifford, S.; Cioslowski, J.; Stefanov, B. B.; Liu, G.; Liashenko, A.; Piskorz, P.; Komaromi, I.; Martin, R. L.; Fox, D. J.; Keith, T.; Al-Laham, M. A.; Peng, C. Y.; Nanayakkara, A.; Challacombe, M.; Gill, P. M. W.; Johnson, B.; Chen, W.; Wong, M. W.; Gonzalez, C.; and Pople, J. A. Gaussian 03, Revision C.02; Gaussian: Wallingford, CT, 2004. S2 Figure S1. ESIMS shows 1:1 DPNDI•ClO4– complex. 30 3 / x 10 M cm 40 50 20 10 0 300 400 500 600 700 800 900 / nm Figure S2. UV/Vis spectra of DPNDI in the absence and presence of Bu4NClO4 shows no CT or ET interactions. Figure S3. 1H NMR (400 MHz, DMF-d6, 25 °C) titration of DPNDI ligand with Bu4NClO4 shows no discernible shift in DPNDI signals, indicating that the electronic properties of the πacidic receptor are not significantly perturbed by weak anion–π interactions, which is consistent with the UV/Vis titration data shown in Figure S2. S3 Cartesian coordinates DPNDI/ClO4N C C C C C C C C C C N C C C C H H H H O O O O C C C N C C H H H H C C C N C C H H H H O O Cl O O 3.01625900 2.40169000 0.92345600 0.19868800 0.86134800 2.34424000 0.26517500 -1.21436400 -1.86940800 -1.13302200 -3.33910800 -4.00991300 -3.40444900 -1.93299100 -1.26001800 0.13837500 0.67409100 -1.83248400 0.84849600 -1.65395300 2.95280300 3.06116000 -4.08865700 -3.97451500 4.46223300 5.15799800 6.55203800 7.25597100 6.55918100 5.16387900 4.63162900 7.13001100 7.14391900 4.64850800 -5.44193500 -5.98850000 -7.37855000 -8.21739400 -7.66933900 -6.29415500 -5.35086200 -7.83873500 -8.36365300 -5.89888600 1.51240300 1.73789500 2.18886700 3.68311200 1.83104500 -1.09456300 -0.45642100 -0.30137900 -0.81883800 -1.46696900 -1.60762200 0.36324600 -0.66453900 0.00853500 0.51808000 0.17878100 -0.34137900 -1.02211400 -1.17505900 -1.81004900 -1.95309800 -2.43018100 -2.18290700 0.77735100 1.04641700 -2.16461900 -0.09002000 -1.45114100 0.72737300 -1.11408600 -2.28755100 -2.24704100 -1.14334200 -0.02698700 0.04913900 -3.20674300 -3.14557200 0.86033100 0.98144600 -0.16258800 0.92595100 1.05277500 0.18833700 -0.85105300 -1.07372700 1.65314200 1.89116000 -1.54266400 -1.93054800 3.44150700 3.72363500 2.93782900 3.04355200 1.47946400 0.29818000 1.39485100 1.33532300 0.23515200 -0.83401500 -0.82301400 2.35549700 0.19262900 1.25582300 2.31549700 1.23132300 0.09751100 -0.98290400 -0.91741600 -1.95236700 -1.91198100 -2.72508400 -2.79481800 3.16981500 3.10648400 -1.72500800 2.35366600 -1.90217700 2.12048100 0.27556700 0.55294100 0.51140300 0.22439400 -0.03716900 -0.03009500 0.78723000 0.71935000 -0.26780000 -0.24841900 0.04195400 -0.63155100 -0.65755200 -0.07030000 0.57374800 0.65939800 -1.12267600 -1.17562600 1.04610600 1.19439800 0.47922900 -1.95105200 -0.76134000 -0.61053100 -0.95372400 Total energy = 2202.6791333 S4