Variation – Chapter 9
advertisement

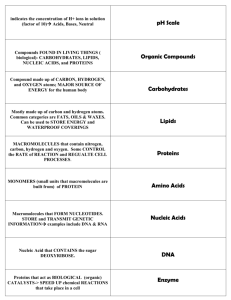
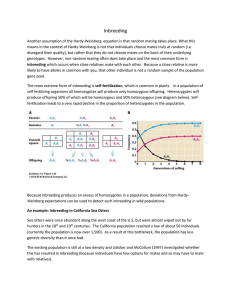
Variation – Chapter 9 Some terms • Genotype • Allele – form of a gene, distinguished by effect on phenotype • Haplotype – form of a gene, distinguished by DNA sequence • Gene copy – number of copies of a given gene, used without distinguishing allele or sequence differences – Allele copies Variation in phenotype can be due to genes AND environment: Is variation genetic, environmental, or both? • Cross phenotypes and use Mendalian ratio predictions • Greater resemblance among siblings than among related individuals points towards genetic contribution • Common garden experiment – See if differences persist over two generations • Genotype frequency is proportion of a genotype A population has a frequency of alleles • Allele frequency is the relative commonness or rarity of an allele Hypothetical population If… • Individuals mate at random and each genotype is equally represented by males and females then.. Hardy Weinberg Equilibrium • After one generation of random mating, genotype frequencies and allele frequencies in a population will remain constant from generation to generation • Has a set of 5 assumptions • H-W can be modified for sex-linked loci and segregation other than 1:1 H-W assumptions • Random mating • Large population • No gene flow • No mutation • Each individual has an equal chance of survival and reproduction – (No natural selection) Genetic Evolution • Asks what happens when one or more of the assumptions are relaxed • Violations of H-W are the major factors the cause evolutionary change Frequencies of alleles • Frequency of heterozygotes are highest when p = q • When an allele is rare, almost all its carriers are heterozygotes – This can cause concealed genetic variation in a population Inbreeding • Inbreeding occurs when individuals are more likely to mate with relatives that with non-relatives or when gene copies are more likely to be identical by descent – Genes are identical by descent if they have descended from a common ancestor relative to other gene copies in the population Inbreeding • As inbreeding proceeds, the frequency of each homozygote increases • Frequency of heterozygotes is H = H0 (1-F) – H0 is frequency of heterozygotes under HW – F is the inbreeding coefficient • F increases as inbreeding continues – F can be calculated by the deficiency of heterozygotes from HW equilibrium Inbreeding • Self-fertilization is the most extreme form of inbreeding Polymorphism • Polymorphism is the presence in a population of two or more variants, either alleles or haplotypes – vs. monomorphism • Can be controlled by one to more than one locus Two loci Genetic variation in viability • Can perform crosses in flies to determine recessive lethal alleles • 10% of alleles on chromosome 2 in Drosophila pseudoobscura are lethal when homozygous • Average person carries 3-5 lethal recessive alleles in heterozygote form • Confirms that there is a great deal of concealed genetic variation Inbreeding depression • Because of these recessive alleles, inbreeding, when increasing the number of homozygotes, reduces survival and fecundity Inbreeding • Inbreeding with selection over many generations can purge deleterious alleles • Many mechanisms for inbreeding avoidance: behavioral and genetic Endangered species Genetic variation in proteins • Enzymes are run through electrophoresis • Electrophoretically distinguishable forms of an enzyme are called allozymes Heterozygosity in proteins • Average heterozygosity was 12% in Drosophila pseudoobscura • Average heterozygosity in humans was 7% Variation in DNA • Variation determined by DNA sequencing • Nucleotide diversity per site (π) • Drosophila melanogaster has 5% average diversity • Humans have .08% average diversity Linkage • Genes are physically associated with other genes on the same chromosome • Changes in allele frequencies at one allele can cause changes in a linked allele Linkage • Two loci in a population are in linkage equilibrium when the genotype of a chromosome at one locus is independent of its genotype at the other locus • Two loci are in linkage disequilibrium when there is nonrandom association between a chromosome’s genotype at one locus and its genotype at the other locus Recombination • Recombination reduces the level of linkage disequilibrium • Tighter linkages take longer to break down from recombination • Linkage disequilibrium is common in asexual populations due to little recombination Determining linkage equilibrium • Use modified HW • Frequency of A1B1/A1B1 in next population should be pA2pB2 if population is in linkage equilibrium Heterostyly in Primula • pin phenotype and thrum phenotype • In most crosses, thrum dominant over pin Quantitative traits • Continuous variation vary because of both polygenic control, as well as environmental factors Genotype x environment interaction • Genotype norm of reaction Causes of variation Estimating variation • Use variance, which measures the spread of values around the mean • Vpheno. = VG + VE • Heritability is the proportion of the phenotypic variance that is genetic – h2 = VG /(VG + VE) Heritability • Two ways to measure – parent-offspring regression – selection experiments Parent-offspring regression • If variation among individuals due to genes, then offspring resemble parents • Plot midparent value against midoffspring value • Slope of line = heritability Parent-offspring regression Selection experiments Selection experiments • Only particular individuals allowed to breed • Difference between mean phenotype of population and mean of selected group = selection differential, S Selection experiments • The change in offspring phenotype between selected group and unselected population is the response to selection, R Heritability estimates