Supplementary Information (doc 1258K)
advertisement

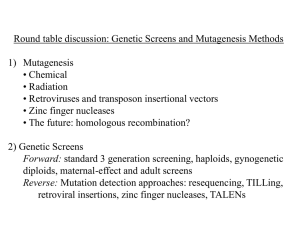
Supplemental Information gene name forward reverse reference Clock TCAGGCCCCACGTCTTTTAC GCCGCCTGGAGGATCAG Kopp et al., 2010 sirt1 ACAGAGCCATGAAGCAGGAT GAGGCACGTCATGAGGAATC this study per1 CCGTCAGTTTCGCTTTTCTC ATGTGCAGGCTGTAGATCCC Cavallari et al., 2011 per2 ATGTCGATGGCTTTAGGCAG CGAGACATCCAGAAGGTGCT Cavallari et al., 2011 cry1a TCCGCTGTGTGTACATCCTC CAAACACTGCAGCAAAAACC Cavallari et al., 2011 cry2a GGACCAATACACCAGCACCAG CAGCAAGTGTCCTGCCATGTC Amaral et al., 2011 rev-erbα TGCTTAGCAGTTATGGCTTTATCG GCAGGGTTTTGACAGAATTTCC this study pparαa ATTATGTACAGCCCTCTGAGCGGA TGAGAACACTTCTGAGGACGGACT Tseng et al., 2011 pparβb CGTCAACACAGCCTACCTGA GCCACAGGGAGTCCATATCA this study pparγ CATACACAAGAAGAGCCGCA ATGTGGTTCACGTCACTGGA this study Agrp GTCCACCTGCAGAGAAGAGG AGAAGGCCTTAAAGAAGCGG Zhang et al., 2010 Npy CCAAACATGAAGATGTGGATGAG CCAAGCAGACGAACAAGAGAAA this study Lep GCAAAATTTACTTCCAA CGCTTTCCCATTTGTTGATT this study Lpl GGCCAAATTTGTCAACTGGT CATGAGCGCCAAGACTGTAA this study SI 1: Primer sequences. Specific forward and reverse primer for qRT-PCR were developed or adopted from literature. SI 2: mRNA expression of hormone and neurotransmitters. (a-c) Relative mRNA expression was quantified by qRT-PCR. Samples were taken at 15 dpf at two different ZT to show alterations in mRNA expression due to circadian rhythms. Values are shown as mean ± SE and significance is marked with *. SI 3: Regression analysis of gene expression of zebrafish larvae raised under control or continuous light (LL) conditions. (a) Ppparβδ and reverbα mRNA expression is highly correlated in zebrafish raised under LL but not control light conditions. (b) Overview of regression analysis of individual genes. Only genes are shown in which regression analysis revealed R² ≥ 0.95. Genes compared are shown with a line. Grey lines indicate a relation under control conditions; black lines indicate a relation under LL conditions. For example, under LL conditions leptin mRNA expression was highly correlated to sirt1, pparβδ, lpl, rev-erbα and per1. SI 4: Adipocytes and lipid profile. (a) Volumes of adipocyte lipid droplets based on methylene signal per fish. Values are shown as mean ± SE. (b) Mean volumes of adipocyte lipid droplets based on methylene signal per fish. Values are shown as mean ± SE. (c) Fatty acid and (d) triglyceride content of pooled and homogenized 15 dpf old larvae zebrafish larvae. Values were normalized to sample DNA content. Values are shown as mean ± SE and significance is marked with *. (e) Lipid profile of 15 dpf old zebrafish larvae under different treatment conditions. Red marks increased values, green marks decreased values [ng / µg DNA]. right eye left eye pineal gland SI 5: Rev-erbα and Pparγ protein expression. (a) Western blot against Rev-erbα (molecular weight: 69.46 kDa) using 100 µg total protein of ten 25 dpf old zebrafish larvae raised under control conditions. (b) Immunohistochemical staining against Rev-erbα (red; Alexa 568 goat anti rabbit) and Pparγ (green; Alexa 488 goat anti mouse) of the zebrafish head region revealed a clear Rev-erbα signal in the pineal gland. (c) Immunohistochemical staining against Rev-erbα and Pparγ of control zebrafish showing the region where adipocytes should develop. No adipocytes and no Pparγ signal were detected. (d) Immunohistochemical staining against Reverbα and Pparγ of control zebrafish showing muscle tissue. Rev-erbα and Pparγ signal colocalized. Marked area was zoomed in (e). SI 6: Survival, activity and size. (a) Survival and (b) activity were monitored over the whole experimental period. (c) DNA content of pooled 15 dpf old zebrafish larvae was quantified by a photospectrometer. SI 7: Transcription Factor Binding Site Analysis (ConTra). Using the online tool ConTra (1) 500 bp promoter regions of zebrafish ppar isoforms (pparα NM_001161333, pparβδ NM_131468 and pparγ NM_131467) were analyzed for Rev-erbα binding sites (REV-RE: A/T GGTCA). Pparα promoter region (500 bp) comprised no REV-REs while pparβδ promoter included three REVREs at position -129, -118 and -77 and pparγ promoter one REV-REs at position -366. No Eboxes for Clock/Bmal binding were found. REV-REs were marked with bold capital letters.