Linkage and Chromosome Mapping in Eukaryotes
advertisement

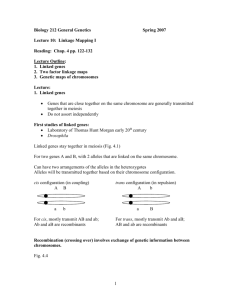
Linkage, Recombination, and Eukaryotic Gene Mapping Genes on the same chromosome are physically and genetically linked They are in the same linkage group Since chromosomes segregate as a unit, linked genes should segregate as a unit Of course, there is recombination (crossing over) that will change that Bateson, Saunders, and Punnett conducted some different crosses with peas They looked at flower color and pollen shape The genes for flower color and pollen shape in peas lie close together on the same chromosome The new combinations of alleles are due to recombinant chromosomes But there are two ways to accomplish this recombination Linkage and recombination To distinguish between linked vs. non-linked genes, we can consider the results of various crosses Depending on the degree of linkage, we may see some characteristic ratios We can also perform a testcross to investigate further Crossing over changes all that The work of Morgan and Sturtevant shed light on the effects of crossing over They looked at crosses involving two X-linked genes at a time The results were unexpected at the time Recombination frequency The percentage of recombinant progeny produced in a cross One map unit (mu) = 1% recombination A different pair of mutations yielded even more confusing results A cross with yellow and miniature gave 35.4% recombinants Sturtevant realized that the recombination frequencies revealed the order of genes and could be used to map them The distance between the genes is crucial when a single crossover occurs Two genes that are more than 50 mu apart behave as if they are on different chromosomes Coupling and repulsion Coupling - when the wild-type alleles are found on one homolog and the mutant alleles are on the other (cis configuration) Repulsion - when one wild-type and one mutant allele are on each homolog (trans configuration) The configuration of the alleles affects the outcome of the testcross Recombination Interchromosomal recombination occurs due to random alignment/segregation of chromosomes in metaphase I/anaphase I Intrachromosomal recombination occurs due to crossing over in prophase I Genetic maps Distances on genetic maps are given in map units (mu, centiMorgans [cM]) 1 mu = 1% recombination Tetrads can also undergo multiple crossovers If the same chromatids are involved, this leads to an exchange of an internal portion of the chromosome The order of genes can be determined by analyzing multiple crossovers Three-point tests Use a testcross for three genes to order the genes in one set of progeny When possible, this is much more efficient than pairwise mapping If the two genes being studied are far apart, the results will tend to underestimate the actual distance between them It doesn’t take into account double crossovers In fact, double crossovers occur at a lower frequency than would be predicted from the individual crossover frequencies The occurrence of one crossover tends to inhibit additional crossovers nearby The degree that one crossover interferes with additional crossovers in the area is interference Physical basis of recombination Exchange between chromosomes is responsible for genetic changes Barbara McClintock and Harriet Creighton were the first to provide concrete evidence for the physical basis of recombination They used a strain of corn with an abnormal chromosome 9 Each end of the abnormal chromosome was easily distinguishable from a normal chromosome 9 That allowed them to clearly show recombinant chromosomes Genomewide association studies These look for non-random associations between a trait and alleles at many different loci Association with a particular haplotype can be used to identify the chromosomal region containing the gene(s) of interest Many haplotypes are based on single-nucleotide polymorphisms (SNPs) Probability of linkage Because the number of human children is small, we need to determine the probability that the genes are linked, using the lod (logarithm of odds) score It is the logarithm of the ratio of the probability of obtaining the results assuming linkage and the probability of obtaining the results assuming independent assortment If the probability for linkage is >1000 times that of the probability for independent assortment (lod score >3), this is convincing evidence for linkage