Supplementary Methods - Word file (33 KB )
advertisement

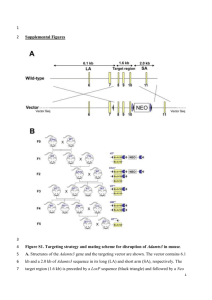
Supplementary Methods Embryo isolation, preparation and screening of single cell cDNAs LS and EB stage 129/SvEv embryos were isolated at E7.0 and E7.5, respectively, in DMEM with 10% fetal calf serum buffered with 25mM HEPES, pH7.4, at room temperature. Preparation and quantification of single cell cDNAs by Southern blotting was performed essentially as described previously1,2 (Fig. 1). For single cell comparisons between mutant and control embryos (Fig. 6), MB to EHF E7.5 embryos were used, where mutant embryos were generated from Blimp-1 (-/-) ES cells (see below), while loxP/loxP ES cell derived or normal wild type embryos were used as controls to isolate single cells. Single cell cDNAs were generated using 30 cycles of PCR-amplification, followed by an additional 30 PCR cycles to determine expression of specific genes. In situ hybridization, antibody staining, and alkaline phosphatase staining In situ hybridization was performed as described previously3. For some experiments, posterior-proximal extraembryonic mesoderm was isolated and disaggregated to determine the number of stained cells. Antibody stainings on dispersed E7.5 germ cell regions and on whole embryos were carried out essentially as described previously 4,5. Antibodies used were: rabbit anti-PGC7/Stella (1:2000, gift from T. Nakano), monoclonal rat anti-GFP (1:500, Nacalai Tesque Inc., Kyoto), rabbit anti-GFP Alexa488 conjugated (1:500, Molecular Probes, Eugene, OR), donkey anti-rabbit IgG Alexa594 conjugated (1:500, Molecular Probes), goat anti-rabbit IgG Alexa488 conjugated (1:500, Molecular Probes) and goat anti-rat IgG Alexa488 conjugated (1:500, Molecular Probes). PGC counts following TNAP-staining was performed as described previously6,7 . BAC modification and transgenic mice production To create Blimp-1mEGFP reporter mice, EGFP coding sequence was subcloned into the BglII and SalI site of the pDisplay vector (Invitrogen). The resulting membranetargeted cassette of EGFP with a polyadenylation signal was amplified and recombined into just after the methyonine of exon 3 of Blimp-1 gene in a 230 kb bacterial artificial chromosome of C57BL/6 backgound bearing 160kb upstream and 70kb downstream region from the Blimp-1 transcription start site purchased from BACPAC Resources Center (http://bacpac.chori.org/), according to the protocol supplied by GeneBridges (http://www.genebridges.com). The modified BAC was injected into male pronuclei of oocytes from C57BL/6/DBA mixed background to generate transgenic mice. Transgenic embryos were imaged on an inverted ZeissLSM 510 confocal microscope. Image-processing and cell counts were performed using Zeiss LSM Image Browser. To create Blimp-1-Cre line, BAC clone #452 P8 (Research Genetics) containing Blimp-1 locus was modified using a PCR-based method8. The coding region in the first exon of Blimp-1 was substituted by the first 100 bp of the Cre sequence preceded by an NLS. The second exon of Blimp-1 was replaced by the remaining Cre sequence followed by a transcriptional STOP cassette. The modified BAC was linearized and microinjected into pronuclei of zygotes derived from B6CBAF1 mice. Generation of Blimp-1 knockout cells, mice and tetraploid chimeras The Blimp-1 targeting vector (pDTA-TK-Blimp-1) was engineered to flank exon 5 with loxP sites and then transfected into E14.1 ES cells (129/ola). Out of 200 ES clones, four clones had one homologously recombined Blimp-1 allele and one clone had both alleles targeted (Clone 2G3). After Cre-mediated deletion of the loxPflanked neor gene, heterozygous mice for the Blimp-1 loxP allele were generated. These mice were bread to deleter-Cre9 to generate the Blimp-1 null allele. The resulting Blimp-1 (+/-) mice where kept on a mixed C57BL/6/CBA/129 background and intercrossed to analyze embryos for PGC-development. To make Blimp-1 (-/-) ES cells, we electroporated double targeted ES cells (clone 2G3) with PGK-Cre plasmid (gift from Sam Aparicio). We identified several -/- clones, of which two were injected into Rosa 26-lacZ10 tetraploid host blastocysts to generate ES cell derived embryos as described previously11. We also got one loxP/loxP clone (2-2A), which was used as a control. References for Supplementary Methods 1. Saito, H., Kubota, M., Roberts, R. W., Chi, Q. & Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679-91 (2004). 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. Saitou, M., Barton, S. C. & Surani, M. A. A molecular programme for the specification of germ cell fate in mice. Nature 418, 293-300 (2002). Wilkinson, D. G. & Nieto, M. A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225, 361-73 (1993). Payer, B. et al. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 13, 2110-7 (2003). Seki, Y. et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278, 440-58 (2005). Cox, W. G. & Singer, V. L. A high-resolution, fluorescence-based method for localization of endogenous alkaline phosphatase activity. J Histochem Cytochem 47, 1443-56 (1999). Lawson, K. A. et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13, 424-36. (1999). Misulovin, Z., Yang, X. W., Yu, W., Heintz, N. & Meffre, E. A rapid method for targeted modification and screening of recombinant bacterial artificial chromosome. J Immunol Methods 257, 99-105 (2001). Schwenk, F., Baron, U. & Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23, 5080-1 (1995). Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70-1 (1999). Nagy, A. Manipulating the mouse embryo : a laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 2003).