Supplementary_Information
advertisement

Supplementary Information for:
On a relationship between molecular polarizability and partial molar
volume in water
Ekaterina L. Ratkovaa and Maxim V. Fedorova,b
a
The Max Planck Institute for Mathematics in the Sciences, Inselstrasse 22,
Leipzig, 04103, Germany, E-mail: fedorov@mis.mpg.de, Tel +49 341 9959
804, Fax +49 341 9959 999
b
Department of Physics, Scottish Universities Physics Alliance (SUPA), Strathclyde
University, John Anderson Building, 107 Rottenrow East, Glasgow, U.K., G4 0NG, email: maxim.fedorov@strath.ac.uk
PART I.
PART II.
PART III.
PART IV
PART V.
PART VI.
Main formulae of the 3D RISM
Details of 3D RISM calculations
Partial Molar Volume (PMV) Estimations
Static Electric Polarizability Estimations
Data for the training set
Data for the test sets
PART I. Main formulae of the 3D RISM
In the three dimensional Reference Interaction Site Model (3D RISM),1 the sixdimensional solute-solvent molecular Ornstein-Zernike (MOZ) equation is
approximated by a set of three-dimensional integral equations via partial
integration over the orientational coordinates. In the case of 3D RISM method
instead of one six-dimensional MOZ equation one has to solve Nsolvent 3D equations.
The equations operate with the intermolecular solvent site - solute total correlation
functions {hα(r)}, and direct correlation functions {cα(r)}:
N
h r c r r r dr
1 R3
(S1)
where ξ, α denote the index of sites in solvent molecule, χξα(r) is the bulk solvent
susceptibility function, and N is the number of sites in a solvent molecule.
The solvent susceptibility function χξα(r) describes the mutual correlations of the
sites of solvent molecules in the bulk solvent. In general, the function can be
obtained from the solvent site-site total correlation functions (hsolvξα(r)) and 3D
structure of a single solvent molecule (intramolecular correlation function ωsolvξα(r):
r solv (r ) hsolv (r )
(S2)
where ρ is the bulk density of the solvent (here and after we imply that each
molecule site is unique in the molecule, so that ρα = ρ for all α).
The solvent susceptibility functions can be calculated one time for a given solvent
at certain thermodynamic conditions and enter the 3D equations as known input
parameters.
To make Eq. (S1) complete, Nsolvent closure relations are introduced:
h (r ) exp u (r ) h (r ) c (r ) B (r ) 1
(S3)
where uα(r) is the 3D interaction potential between the solute molecule and α site of
solvent, Bα(r) are bridge functions, β = 1/kBT, kB is the Boltzmann constant, and T is
the temperature.
The 3D interaction potential between the solute molecule and α site of solvent
(uα(r), Eq. (S3)) is estimated as a superposition of the site-site interaction potentials
between solute sites and the particular solvent site (usα(r), where index s denotes
the site in a solute molecule and index α - the site in a solvent molecule), which
depend only on the absolute distance between the two sites:
N
u (r ) u s rs r
(S4)
s 1
where rs is the radius-vector of solute site (atom).
We used the common form of the interaction potential represented by the longrange electrostatic term uelsα(r) (where r = |rs−r|) and short-range Lennard-Jones
(LJ) term uLJsα(r) as:
us (r ) usel (r ) usLJ (r ),
qs q
,
r
LJ
LJ
LJ s
us (r ) 4 s
r
usel (r )
r
12
LJ
s
(S5)
6
where {qs, qα} are the partial electrostatic charges of the corresponding solute and
solvent sites, and {εLJsα,σLJsα} are the LJ solute-solvent interaction parameters.
In general, the bridge functions Bα(r) in Eq. (S3) can be written as an infinite series
of integrals over high order correlation functions and are therefore practically
incomputable. Thus, some approximations are introduced. The most
straightforward and widely used model is the RISM/HNC approximation, which
sets Bα(r) to zero. However, due to the uncontrolled growth of the argument of the
exponent the use of the HNC closure can lead to divergence of the numerical
solution of the RISM equations in some cases. One way to overcome this problem
is to linearize the exponential function for arguments larger than certain constant C:
exp (r ) 1, (r ) C
h (r )
(
r
)
exp
C
C
1
,
(
r
)C
(S6)
where Ξα(r) = −βuα(r) + hα(r) − cα(r). The partially linearized HNC closure for the
case C = 0 was proposed by Kovalenko and Hirata2 and named as KH closure. In
the current work we performed calculations with the KH closure. We note that in
the literature the combination of the KH closure and the 3D RISM equations are
usually referred to as 3D RISM-KH theory.
PART II. Details of the 3D RISM calculations
The 3D RISM calculations were performed using the NAB simulation package 3 in
the AmberTools 1.4 set of routines.4 The 3D-grid around a solute was generated
such that the minimal distance between any solute atom and the edge of solvent
box (buffer in NAB notation) was equal to 30 Å, whereas the linear grid spacing in
each of the three directions was 0.3 Å. We employed the MDIIS iterative scheme,5
where we used 5 MDIIS vectors, MDIIS step size - 0.7, and residual tolerance is
10-10. The KH closure was used for solution of the 3D RISM equations.
The solvent susceptibility functions for 3D RISM calculations were obtained by the
1D RISM method present in the AmberTools 1.4. The dielectrically consistent 1D
RISM (DRISM)6 was employed with the KH closure. The grid size for 1Dfunctions was 0.025 Å, which gave a total of 16384 grid points. We employed the
MDIIS iterative scheme, where we used 20 MDIIS vectors, MDIIS step size - 0.3,
and residual tolerance - 10-12. The solvent was considered to be pure water with the
number density 0.0333 Å-3, a dielectric constant of 78.497, at a temperature of
300K. The final susceptibility solvent site-site functions were stored and then used
as input for the 3D RISM calculations.
Within the 3D RISM approach we perform calculations with the following solutes
parameters:
(1) Coordinates of each molecule were optimized using the AM1 Hamiltonian 7 via
the antechamber8 suite, which uses the sqm4 program for semiempirical QM
calculations. The initial configurations for these QM geometry optimizations were
taken from the previous 1D RISM calculations.9
(2) Atomic partial charges were calculated using the AM1-BCC method8,10
implemented in the antechamber from the AmberTools 1.4 package.4
(3) The LJ parameters from the General Amber Force Field (GAFF) 8 were assigned
to solute atoms with the antechamber and the tleap programs.8 In the case of 1D
RISM calculations, for all atoms with zero GAFF LJ potential parameters the
following parameters were used σLJ =0.4 Å and εLJ = 0.1185 kcal/mol to prevent
divergence of the algorithm.
PART III. Partial Molar Volume (PMV) Estimations
In this work, PMV values were calculated as follows. Direct correlation
functions of the solute – solvent systems were obtained by 3D RISM method
combined with the Kovalenko-Hirata closure (3D RISM-KH). The resulted
correlation functions were used to calculate the PMV values by the following
Kirkwood-Buff equation:
N
V = k BT T 1 3c (r )dr ,
R
=1
(S7)
where V is the partial molar volume, kB is the Boltzmann constant, T is the
temperature, χT is the pure solvent isothermal compressibility, ρ is the bulk density
of the solvent, cα(r) is the intermolecular solute-solvent site direct correlation
function (α denote the index of the corresponding solvent molecule site), N is the
overall number of the solvent sites in one solvent molecule, r is the radius-vector of
the solvent site. We note that previously this formula was referred as the 3D RISMKH/KB theory.11
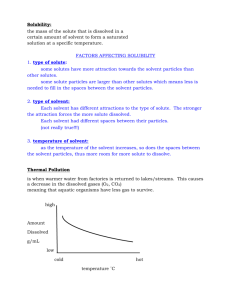
We found a strong linear correlation between the calculated and experimental
PMVs12-16 with the correlation coefficient r = 0.987 and standard deviation of error
of 3.98 cm3mol-1 (6.61 Å3) which is about 4% of the average value of PMV. One
can see that the calculated values have a bias from the corresponding experimental
data, which can be eliminated with two empirical coefficients:
V a1 V 3D RISM KH / KB a0 ,
(S8)
where V 3D RISM KH / KB is the PMV calculated with Eq. (S7), a1 = 1.04 is the scaling
coefficient, a0 = 1.63 cm3mol-1 (2.71 Å3) is the intercept. The fact that the PMV
obtained by the 3D RISM-KH/KB method is so well correlated with the
corresponding experimental values allows us to use them as an accurate estimation
of experimental data.
We note that the literature data of PMV are published in cm3mol-1. However,
to make units of PMV and polarizability consistent we used Å3 instead of cm3mol-1
in the main text of the article.
Figure S1: Correlations between calculated and experimental values of partial
molar volume in water (V) , where calculated values are obtained by the 3D RISMKB method with AmberTools 1.4. Solid black lines give the line-of-best-fit while
the dotted black line illustrates the ideal correlation; r is the correlation coefficient
between averaged experimental and corresponded calculated values.
Table S1. Experimental and calculated data for partial molar volume in water (cm3mol-1), where calculated values
are obtained by the 3D RISM-KB method.
Experimental data
Name
ethane
methane
propane
buta-1,3-diene
ethylbenzene
n-propylbenzene
toluene
2-methylbutan-2-ol
2-methylpropan-1-ol
butan-1-ol
butan-2-ol
ethanol
heptan-1-ol
hexan-1-ol
hexan-3-ol
methanol
pentan-1-ol
pentan-2-ol
pentan-3-ol
propan-1-ol
propan-2-ol
Ref. [12] Ref. [13] Ref. [14]
51.20
52.00
37.30
37.30
67.00
69.00
68.30
114.50
97.71
102.50
86.75
86.48
86.65
55.12
133.43
117.56
117.14
38.25
102.88
102.55
101.28
70.63
71.93
86.60
55.10
118.70
Ref. [15]
Ref. [16]
114.75
130.75
98.43
97.50
101.30
86.50
86.60
86.60
55.10
133.00
118.50
117.14
38.15
102.40
38.20
102.40
101.20
70.70
71.80
101.20
70.60
71.95
86.70
86.60
86.60
55.10
mean
value
51.60
37.30
68.00
68.30
114.63
130.75
97.88
101.90
86.65
86.57
86.62
55.11
133.22
118.25
117.14
38.20
102.56
102.55
101.23
70.64
71.89
3D RISM-KB
48.45
33.28
63.33
66.09
106.18
121.14
91.00
94.27
79.26
79.95
79.99
50.04
124.93
109.96
109.92
34.29
94.95
94.97
94.87
65.03
65.32
acetaldehyde
3-methylbutan-2-one
95.00
4-methylpentan-2-one
95.00
butanone
82.90
pentan-2-one
98.00
pentan-3-one
98.08
propanone
66.80
di-n-propyl ether
115.00
diethyl ether
90.40
diisopropyl ether
1,2-dimethoxyethane
95.88
2-butoxyethanol
122.91
2-ethoxyethanol
90.97
2-propoxyethanol
107.10
3-hydroxybenzaldehyde
4-hydroxybenzaldehyde
dimethoxymethane
43.70
82.50
98.00
82.90
98.00
90.40
90.40
115.00
95.70
97.90
96.90
80.50
43.70
95.00
95.00
82.77
98.00
98.08
66.80
115.00
90.40
115.00
95.79
122.91
90.97
107.10
97.90
96.90
80.50
46.92
91.59
106.50
77.43
92.45
92.26
62.62
112.44
82.34
112.20
84.66
113.94
83.88
98.94
91.67
91.56
70.41
PART IV. Static Electric Polarizability Estimations
In the case of the static electric polarizability, we performed calculations with the
Gaussian03 software17 at B3LYP/aug-cc-pVDZ level of theory. This approach was
found to provide with one of the most reliable data of static electric polarizability
according to the analysis performed by the National Institute of Standards and
Technology (NIST).18 Indeed, we obtained a good agreement between the
calculated static electric polarizabilities and the corresponding experimental
data18,19 (r = 0.997, standard deviation of error equals 0.33 Å3 (≈ 3% of the average
value of polarizability), see Figure S2). For the rest of the work we used the
calculated values of polarizability as a precise estimation of experimental data.
Figure S2: Correlations between the calculated and experimental values of static
electric polarizability (α), where calculated values are dipole electric field
polarizabilities computed at B3LYP/aug-cc-pVDZ level of theory with Gaussian 03
software; solid black lines give the line-of-best-fit; r is the correlation coefficient
between averaged experimental and corresponded calculated values.
Example of the Guassian03 input file:
%chk =input.chk
# polar b3lyp/aug-cc-pVDZ
methane
01
C -0.445783 -0.012048 0.000000
H -0.082284 -1.040219 -0.000001
H -0.082264 0.502030 0.890418
H -0.082266 0.502031 -0.890418
H -1.536318 -0.012035 0.000001
1
2
3
4
5
2345
1
1
1
1
Table S2. Experimental and calculated data for static electric polarizability (Å3),
where calculated values are obtained at B3LYP/aug-cc-pVDZ level of theory.
Name
2,2,4-trimethylpentane
n-decane
n-hexane
n-octane
n-pentane
propane
hept-1-ene
hex-1-ene
ethanol
chloroethane
acetaldehyde
butyraldehyde
propionaldehyde
butanone
pentan-2-one
pentan-3-one
propanone
di-n-propyl_ether
diethyl_ether
ethane
methane
n-butane
n-heptane
n-nonane
pent-1-ene
1,3,5-trimethylbenzene
m-xylene
o-xylene
p-xylene
methanol
propan-1-ol
propan-2-ol
1,2-dichlorobenzene
Experimental data
Ref. [18]
Ref. [19]
15.44
19.15
11.45
11.81
15.48
9.58
9.99
5.92
6.29
13.51
11.65
5.11
5.01
6.40
4.28
4.59
8.18
6.35
8.16
9.93
9.93
6.10
12.53
8.73
4.23
4.47
2.45
2.60
7.69
8.12
13.65
17.36
9.65
15.76
14.21
14.15
14.11
3.08
6.77
6.97
14.20
calculated
15.15
19.32
11.70
15.45
9.82
6.15
13.70
11.79
5.02
6.20
4.57
8.15
6.31
8.11
9.98
9.87
6.36
12.56
8.86
4.31
2.52
7.94
13.58
17.36
9.89
16.52
14.46
14.35
14.50
3.16
6.82
6.86
14.23
1,3-dichlorobenzene
1,4-dichlorobenzene
2-methylstyrene
chlorobenzene
penta-1,4-diene
14.28
14.20
16.05
12.25
11.49
14.42
14.50
16.68
12.38
9.97
PART V. Data for training set
Table S3. Composition of training set. Data for static electric polarizability (α) and partial molar volume in water
( V ), where benchmark values of the partial molar volume are obtained with Eq. (8).
Name
Alkanes
ethane
methane
n-butane
n-decane
n-heptane
n-hexane
n-nonane
n-octane
n-pentane
propane
Alkenes
ethene
hept-1-ene
hex-1-ene
non-1-ene
oct-1-ene
Alkylbenzenes
ethylbenzene
isobutylbenzene
α (Å3)
(benchmark)
No. of
π-electrons
No. of
lone pairs
4.31
2.52
7.93
19.32
13.58
11.70
17.36
15.45
9.82
6.15
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
86.69
60.39
137.04
293.60
215.42
189.56
267.05
240.95
163.46
112.49
86.59
61.75
136.77
294.47
214.97
188.92
267.35
240.91
162.85
112.09
4.16
13.70
11.79
17.53
15.61
2
2
2
2
2
0
0
0
0
0
75.81
206.05
180.05
258.13
232.14
70.19
202.44
175.95
255.51
228.90
14.31
17.99
6
6
0
0
186.78
237.43
182.34
233.36
V (Å3)
benchmark
predicted
m-xylene
n-butylbenzene
n-hexylbenzene
Alcohols
butan-1-ol
butan-2-ol
decan-1-ol
ethanol
heptan-1-ol
hexan-1-ol
hexan-3-ol
methanol
nonan-1-ol
octan-1-ol
Phenols
4-n-propylphenol
4-tert-butylphenol
o-cresol
p-cresol
phenol
1-Chloroalkanes
1-chlorobutane
1-chloroheptane
1-chlorohexane
1-chloropentane
1-chloropropane
Aldehydes
acetaldehyde
14.46
18.20
22.08
6
6
6
0
0
0
186.88
238.69
290.61
184.51
236.23
290.04
8.67
8.67
20.07
5.02
14.33
12.43
12.34
3.16
18.15
16.23
0
0
0
0
0
0
0
0
0
0
2
2
2
2
2
2
2
2
2
2
141.30
141.37
297.41
89.45
219.29
193.33
193.26
62.14
271.27
245.35
141.21
141.19
299.12
90.68
219.58
193.28
192.05
64.96
272.53
246.01
17.15
18.76
13.15
13.24
11.20
6
6
6
6
6
2
2
2
2
2
217.24
237.18
164.95
164.92
138.53
216.02
238.29
160.62
161.87
133.60
9.97
15.71
13.79
11.88
8.08
0
0
0
0
0
3
3
3
3
3
155.12
233.00
207.10
181.16
129.25
156.38
235.86
209.31
182.89
130.24
4.57
0
2
84.04
84.48
butyraldehyde
formaldehyde
heptanal
Ketones
hexan-2-one
nonan-2-one
octan-2-one
pentan-2-one
Ethers
di-n-butyl ether
di-n-propyl_ether
diethyl_ether
diisopropyl_ether
dimethyl_ether
Dienes
2,3-dimethylbuta-1,3-diene
2-methylbuta-1,3-diene
buta-1,3-diene
Styrenes
2-methylstyrene
2-ethylstyrene
tert-butylstyrene
Polychlorinated alkanes
1,4-dichloropentane
dichloromethane
pentachloroethane
tetrachloromethane
trichloromethane
8.15
2.64
13.80
0
0
0
2
2
2
135.60
56.88
213.66
134.05
57.65
212.27
11.86
17.58
15.66
9.98
0
0
0
0
2
2
2
2
188.91
266.95
240.89
162.98
185.44
264.63
238.12
159.32
16.38
12.56
8.86
12.33
5.08
0
0
0
0
0
2
2
2
2
2
249.56
197.64
145.44
197.22
90.80
248.06
195.15
143.82
191.99
91.46
11.91
10.26
8.57
4
4
4
0
0
0
165.11
141.62
117.27
163.31
140.48
117.14
16.68
18.54
22.84
8
8
8
0
0
0
199.72
225.68
272.90
201.01
226.69
286.34
13.76
6.24
13.82
10.17
8.16
0
0
0
0
0
6
6
15
12
9
197.69
94.45
177.63
136.45
113.91
200.32
96.18
175.34
133.43
114.19
Polychlorinated alkenes
cis-1,2-dichloroethene
tetrachloroethene
trans-1,2-dichloroethene
trichloroethene
Polychlorinated benzenes
1,2,3,4-tetrachlorobenzene
1,3,5-trichlorobenzene
1,3-dichlorobenzene
1,4-dichlorobenzene
chlorobenzene
2-chlorotoluene
7.98
12.19
8.20
10.13
2
2
2
2
6
12
6
9
112.41
151.98
112.80
132.48
106.03
147.09
109.02
127.18
19.01
16.98
14.42
14.50
12.38
14.46
6
6
6
6
6
6
12
9
6
6
3
3
209.53
191.50
170.09
170.49
152.38
178.51
213.22
193.66
166.67
167.82
147.09
175.89
PART VI. Data for the test sets
Table S4. Composition of the internal and external test sets. Data for static electric polarizability (α) and partial
molar volume in water ( V ), where benchmark values of the partial molar volume are obtained with Eq. (8).
Name
Internal test set
2,2,4-trimethylpentane
2,2,5-trimethylhexane
2,2-dimethylbutane
2,2-dimethylpentane
2,3,4-trimethylpentane
2,3-dimethylpentane
2,4-dimethylpentane
2-methylbutane
2-methylhexane
2-methylpentane
3-methylhexane
3-methylpentane
2-methylbut-2-ene
2-butene-1,4-diol
3-methylbut-1-ene
but-1-ene
pent-1-ene
propene
trans-hept-2-ene
α (Å3)
(benchmark)
No. of
π-electrons
No. of
lone pairs
15.15
17.04
11.51
13.39
15.06
13.31
13.40
9.77
13.53
11.64
13.45
11.56
9.74
9.58
9.86
8.00
9.89
6.09
13.82
0
0
0
0
0
0
0
0
0
0
0
0
2
2
2
2
2
2
2
0
0
0
0
0
0
0
0
0
0
0
0
0
4
0
0
0
0
0
V (Å3)
benchmark
predicted
233.37
260.53
184.93
210.86
231.63
209.32
211.53
162.09
214.05
188.06
212.75
186.89
153.24
134.46
153.91
128.26
154.15
101.95
206.44
236.74
262.94
186.31
212.39
235.42
211.30
212.47
162.19
214.26
188.16
213.23
187.01
147.60
133.92
149.22
123.45
149.63
96.99
204.12
1,2,3-trimethylbenzene
1,2,4-trimethylbenzene
1,3,5-trimethylbenzene
2-ethyltoluene
4-ethyltoluene
n-pentylbenzene
n-propylbenzene
o-xylene
p-xylene
sec-butylbenzene
tert-butylbenzene
toluene
2-methylbutan-1-ol
2-methylbutan-2-ol
2-methylpentan-2-ol
2-methylpentan-3-ol
2-methylpropan-1-ol
3-methylbutan-1-ol
4-methylpentan-2-ol
pentan-1-ol
pentan-2-ol
pentan-3-ol
propan-1-ol
propan-2-ol
2,3-dimethylphenol
2,4-dimethylphenol
2,5-dimethylphenol
2,6-dimethylphenol
16.28
16.45
16.52
16.18
16.43
20.16
16.28
14.35
14.50
18.01
17.87
12.41
10.38
10.37
12.26
12.21
8.60
10.45
12.31
10.54
10.54
10.44
6.82
6.86
15.07
15.21
15.30
14.92
6
6
6
6
6
6
6
6
6
6
6
6
0
0
0
0
0
0
0
0
0
0
0
0
6
6
6
6
0
0
0
0
0
0
0
0
0
0
0
0
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
208.70
211.36
213.33
208.67
213.40
264.77
212.72
184.66
186.93
235.86
232.52
160.47
164.86
166.14
192.22
190.77
140.11
165.45
191.55
167.31
167.34
167.17
115.44
115.95
189.39
191.51
190.74
188.46
209.68
212.07
213.05
208.27
211.80
263.37
209.70
182.87
185.03
233.58
231.69
156.08
164.89
164.82
190.96
190.26
140.33
165.83
191.66
167.16
167.19
165.75
115.61
116.18
187.19
189.10
190.31
185.12
3,4-dimethylphenol
3,5-dimethylphenol
3-ethylphenol
4-ethylphenol
2-chloro-2-methylpropane
2-chlorobutane
2-chloropropane
chloroethane
chloromethane
hexanal
isobutyraldehyde
nonanal
octanal
pentanal
propionaldehyde
3-methylbutan-2-one
4-methylpentan-2-one
butanone
decan-2-one
heptan-2-one
pentan-3-one
propanone
undecan-2-one
methyl tert-butylether
methylethyl ether
1,1,1,2-tetrachloroethane
1,1,1-trichloroethane
1,1,2,2-tetrachloroethane
15.19
15.30
15.15
15.18
9.88
9.86
8.07
6.20
4.32
11.90
8.11
17.62
15.70
10.02
6.31
9.86
11.76
8.11
19.49
13.76
9.87
6.36
21.42
10.38
6.96
11.96
10.07
12.02
6
6
6
6
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
2
2
2
2
3
3
3
3
3
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
2
12
9
12
189.30
191.53
191.43
191.45
155.07
154.92
129.60
103.40
76.74
187.58
135.36
265.58
239.70
161.63
109.73
161.49
187.33
136.93
292.92
214.91
162.64
111.25
318.98
168.57
118.19
159.20
142.62
157.07
188.91
190.33
188.26
188.67
155.15
154.90
130.08
104.17
78.11
186.01
133.47
265.16
238.68
159.90
108.51
157.72
184.02
133.49
291.20
211.72
157.90
109.18
317.85
164.93
117.60
158.23
140.59
159.01
1,1,2-trichloroethane
1,1-dichloroethane
1,2-dichloroethane
1,2-dichloropropane
1,3-dichloropropane
1,2,3,5-tetrachlorobenzene
1,2,3-trichlorobenzene
1,2,4,5-tetrachlorobenzene
1,2,4-trichlorobenzene
1,2-dichlorobenzene
1,1-dichloroethene
1,1,2-trichloropropene
1,1,3-trichloropropene
1,1-dichlorobut-1-ene
1,4-dichlorobut-2-ene
hexa-1,5-diene
penta-1,4-diene
External test set: druglike molecules
2-acetaminophen
2-hydroxybenzoic acid
2-methyoxybenzoic acid
3-acetaminophen
3-hydroxybenzoic acid
3-methoxybenzoic acid
4-hydroxybenzoic acid
4-methoxybenzoic acid
acetanilide
aspirin
9.95
8.18
8.27
9.93
10.06
18.91
16.60
19.12
16.99
14.23
8.03
12.06
12.24
11.90
12.34
11.81
9.97
0
0
0
0
0
6
6
6
6
6
2
2
2
2
2
4
4
9
6
6
6
6
12
9
12
9
6
6
9
9
6
6
0
0
138.82
122.30
120.12
146.35
145.79
208.82
189.64
209.55
191.59
169.20
114.23
157.90
157.04
166.04
162.23
169.82
143.98
138.91
123.04
124.23
147.18
149.07
211.85
188.36
214.66
193.74
164.13
106.68
153.91
156.50
160.25
166.41
161.92
136.42
16.95
14.05
16.11
17.01
13.99
16.03
14.20
16.39
16.26
17.67
6
6
6
6
6
6
6
6
6
6
5
6
6
5
6
6
6
6
3
8
196.59
164.13
191.30
196.04
163.37
191.20
163.29
191.52
191.91
220.59
204.66
161.60
190.14
205.51
160.76
189.01
163.70
194.00
200.78
206.04
benzoic acid
13.14
butylparaben
21.81
ethylparaben
18.10
ibuprofen
24.41
methylparaben
16.18
paracetamol
17.12
phenacetin
21.41
propylparaben
19.90
triclocarban
33.18
External test set: other polyfunctional molecules
2-butoxyethanol
13.16
2-chlorophenol
13.23
2-ethoxyethanol
9.42
2-methoxyphenol
13.92
2-phenylethanol
15.02
2-propoxyethanol
11.26
2,4-hexadienal
14.32
2-ethyl-4-methylhexa-2,4-dienal
18.35
3-chlorophenol
13.27
3-hydroxybenzaldehyde
13.86
3-methoxyphenol
14.07
3-phenylpropanol
16.98
4-chloro-3-methylphenol
15.15
4-chlorophenol
13.37
4-hydroxybenzaldehyde
14.11
4-methoxyacetophenone
18.07
E-but-2-enal
8.47
E-hex-2-enal
12.42
6
6
6
6
6
6
6
6
12
4
6
6
4
6
5
5
6
13
159.23
273.14
221.07
315.55
192.88
196.32
251.51
247.24
335.83
154.76
269.11
217.76
310.80
191.15
207.02
266.40
242.68
363.84
0
6
0
6
6
0
4
4
6
6
6
6
6
6
6
6
2
2
4
5
4
4
2
4
2
2
5
4
4
2
5
5
4
4
2
2
200.24
156.19
148.12
171.99
189.39
174.21
167.21
243.64
156.54
161.62
170.90
215.53
182.01
157.67
161.44
215.21
125.69
177.90
197.66
153.11
145.84
165.54
186.43
171.34
191.06
246.87
153.70
164.67
167.59
213.61
179.78
155.10
168.21
223.01
124.20
178.96
E-oct-2-enal
acetophenone
allyl_alcohol
benzyl_alcohol
ethyl_phenyl_ether
methyl_phenyl_ether
16.33
14.74
6.84
13.14
15.07
13.07
2
6
2
6
6
6
2
2
2
2
2
2
229.80
182.95
104.74
162.06
196.53
168.85
233.12
182.65
101.59
160.48
187.19
159.41
References:
(1) Hirata, F. (ed.) Molecular theory of solvation Kluwer Academic Publishers,
Dordrecht, Netherlands, 2003
(2) Kovalenko, A. and Hirata, F. J. Phys. Chem. B 1999, 103, 7942-7957.
(3) Luchko, T.; Gusarov, S.; Roe, D. R.; Simmerling, C.; Case, D. A.;
Tuszynski, J. and Kovalenko, A. J. Chem. Theory Comput. 2010, 6, 607-624.
(4) Case, D. A.; Cheatham, T. E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.
M.; Onufriev, A.; Simmerling, C.; Wang, B. & Woods, R. J. Comput. Chem.
2005, 26, 1668-1688.
(5) Kovalenko, A.; Ten-No, S. and Hirata, F. J. Comput. Chem. 1999, 20, 928936.
(6) Perkyns, J. S. and Pettitt, B. M. Chem. Phys. Lett. 1992, 190, 626-630.
(7) Dewar, M. J. S.; Zoebisch, E. G.; Healy, E. F. and Stewart, J. J. P. J. Am.
Chem. Soc. 1985, 107, 3902-3909.
(8) Wang, Z. X.; Zhang, W.; Wu, C.; Lei, H. X.; Cieplak, P. and Duan, Y. J.
Comput. Chem. 2006, 27, 994-994.
(9) Ratkova, E. L.; Chuev, G. N.; Sergiievskyi, V. P. and Fedorov, M. V. J.
Phys. Chem. B 2010, 114, 12068-12079.
(10) Jakalian, A.; Bush, B. L.; Jack, D. B. and Bayly, C. I. J. Comput. Chem.
2000, 21, 132-146; Jakalian, A.; Jack, D. B. and Bayly, C. I. J. Comput.
Chem. 2002, 23, 1623-1641.
(11) Imai, T.; Harano, Y.; Kovalenko, A. and Hirata, F. Biopolymers 2001, 59,
512-519; Imai, T.; Ohyama, S.; Kovalenko, A. and Hirata, F. Protein Sci.
2007, 16, 1927-1933.
(12) S. Cabani; et al. J. Solution Chem. 1981, 10, 563.
(13) H. Durchschlag; et al. Radiat. Phys. Chem. 2003, 67, 479.
(14) A.V. Plyasunov; E.L. Shock. Geochimi. Cosmochim. Acta 2000, 64, 439.
(15) J.T. Edward; et al. J. Chem. Soc. Faraday Trans. I 1977, 73, 705.
(16) S. Sawamura; et al. J. Phys. Chem. B 2001, 105, 2429.
(17) Frisch, et al. Gaussian 03 Gaussian, Inc., 2004
(18) http://cccbdb.nist.gov
(19) K. Miller. J. Am. Chem. Soc. 1990, 112, 8533