Methods Statistical analysis Gene-by
advertisement

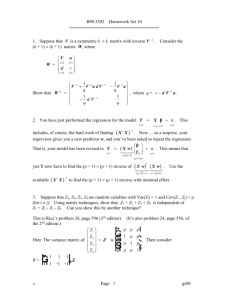
Methods Statistical analysis Gene-by-environment interaction analysis The general test for GxE interaction in sib pair-based association analysis of quantitative traits (van der Sluis et al., 2008) is an extension of the Fulker et al. (1999) maximum likelihood variance components analysis of quantitative traits in sib-pairs data that incorporates environmental main effects plus GxE effects (van der Sluis et al., 2008). Since the association effect is decomposed here into within-family (w) and between-family (b) effects, the design is robust against spurious association stemming from population stratification (van der Sluis et al., 2008). As the association between the GxE interaction and a phenotype is also susceptible to population stratification bias, the sib pair design allows for extending the orthogonal decomposition into between- and within-family effects also to the GxE interaction effects. Here, to partition the additive QTL effects into b and w effects and to model the GxE effects, we followed the model described by van der Sluis et al. (2008), except that we included only the additive effects, and adopted the Abecasis et al. (Abecasis et al., 2000a,b) genotype coding to make use of parental genotypes and to handle sibships of any size, with or without parental genotypes (Mascheretti et al., 2013). Briefly, if we assume that (1) a diallelic marker has allele A1 with frequency p, and allele A2 with frequency 1 – p = q, and genotypes A1A1, A1A2 and A2A2 with genotypic effects a, 0 and –a, respectively, under an additive model, (2) the observed trait value of an individual is a function of a major gene effect (QTL), an additive polygenic genetic background effect, a shared environmental effect, and a non-shared environmental effect (which includes measurement error), (3) the effects of the additive polygenic genetic background, the common and the non-shared environment, and the QTL are mutually uncorrelated, and (4) the additive polygenic genetic background effect and the environmental effects are normally distributed with mean = 0, the additive QTL effects can then be orthogonally partitioned into b and w effects, as specified in Abecasis et al. (Abecasis et al., 2000a,b). Abecasis et al. (2000a,b) extended the Fulker et al. (1999) model to accommodate any number of offspring, with or without parental genotypes, as follows: ∑jgij if parental genotypes are unknown ni bi = , and: giF + giM if parental genotypes giF and giM are available 2 wij = gij -bi , so that bi is the expectation of each sib genotype gij conditional on family i genotype data and wij is the deviation from this expectation for offspring j. Significant positive values of the within-pairs component indicate that a child inherits more copies of the investigated allele than would be expected. In order to partition the additive QTL effects into between- and within-family effects (Mascheretti et al., 2013), we tested the rare allele of each SNPs against the major allele (see Genotyping section). More recently, to model the interaction effect, van der Sluis et al. (2008) adopted the notation proposed by van den Oord (1999) in which the environmental main effect (e) with a dichotomous environmental exposure is modelled as the difference in the phenotypic means of environmental Conditions 1 and 2, and the interaction effect is such that subjects in Condition 2 with genotypes A1A1, A1A2 and A2A2 have effects –i, 0 and i, respectively. Under this model, the interaction parameter i represents the difference between genotypic value a in Condition 2, and genotypic value a in Condition 1 after the main effect of the environmental condition has been taken into account. Differently from van der Sluis et al. (2008), however, we did not decompose the environmental variables that vary from one sibling to the next within the same sibship into the b and w parameters. Instead, we limited ourselves to use the sibling specific value, because the performance of a child in the neuropsychological domains depended only on his/her environmental condition and not on the environmental condition of his/her siblings. In the case of the sib pair association design the phenotypic score yijkg (i.e., the observed score y of subject j from family i in condition k with genotype g) is then modelled as: yijkg = τi + abAbi + awAwij + ekEk + ibkgIkg + iwkgIkg + εij where τi is the family-specific intercept, ab and aw are the estimated between- and within-family additive genetic effects of the marker weighted by the derived coefficients Abi and Awij (where bi and wij are orthogonal between- and within-family components of the genotype gij), ek represents the effects of the categorical environmental condition k, ibkg and iwkg represent the between- and within-family effects of the interaction of genotype g and environmental condition k, weighted by the derived coefficient Ikg, and εij is the residual term (van der Sluis et al., 2008). Since each quantitative environmental variable was centred around its mean, the environmental main effect (e) was then modelled as just described for the categorical environmental variables. In the case of the sib pair association design, the phenotypic score yijg (i.e., the observed score y of subject j from family i with genotype g) was then modelled as: yijg = τi + abAbi + awAwij + eEij + ibgEijAbi + iwgEijAwij +εij where e represents the effect of an increase of one unit of the quantitative environmental exposure, ibg and iwg represent the between- and within-family effects of the interaction of genotype g and the environmental measure Eij. Our method of ascertainment resulted into a left-skewed distribution of the association model residuals. To obtain valid p-values in the presence of departures from normality of the residuals, pvalues were computed by applying a permutation procedure to the residuals of a model without the within-family interaction term iw. For instance the model for categorical covariates becomes: Yijkg = τi + abAbi + awAwij + ekEk + ibkgIkg + εij Prior to the permutations, we imputed the values of the phenotypes and quantitative environmental variables that were missing in the dataset using an EM algorithm implemented in the Missing Value Analysis (MVA) function of SPSS version 17.0. Few values were missing for any variable (on average 10%), and the imputation had therefore little impact on the coefficient and variance component estimates and their precision in the actual data. We implemented the permutation procedure using the R statistical environment (www.rproject.org). Permutations were applied to the sibships, to preserve the within-sibship phenotype correlation. The varying sibship size required the following adjustments. Let mi* be the size of the permuted sibship providing the residuals and mi the size of the sibship providing the fitted values. When mi* ≥ mi, mi residuals were randomly sampled without replacement from the mi* available residuals and added to the mi fitted values. When mi* < mi, mi residuals were sampled with replacement from the mi* available residuals, so that at least one residual was used two or more times. The permutation procedure was repeated 1.000 times for each analysis. Since Bonferroni correction for multiple testing would have been too conservative (Scerri and Schulte-Korne, 2010), we decided to adjust the significance levels by the false discovery rate (FDR) method (Storey, 2002) applied to the 28 tests performed for each marker (7 environmental variable x 4 phenotypes), separately for each marker (Mascheretti et al., 2013). Gender was taken into account in the extended equation because probands’ sex ratio (males:females) in our sample was nearly 3:1, and it may imply differences in mean scores between males and females. Moreover, since simulations studies revealed specific situations (e.g., irregular distributions of the variables, ease of analysis, prior use of the variable) in which dichotomized variables performed as well as or better than the original quantitative factors, we subsequently decided to further explore this dataset by conducting GxE analysis with dichotomized environmental factors, which is indeed more straightforward than conducting analyses with continuous indicators, especially when testing interaction effects in regression (DeCoster et al., 2009). We therefore decided to dichotomize raw scores of quantitative environmental variables (i.e., birth weight, parental age, SES, and parental education; see ‘Methods’, ‘Environmental data collection’ section). In particular, birth weight and SES have been dichotomized based on theoretically meaningful cut-off points, as available from existent literature (Zubrick et al., 2007; Lasky-Su et al., 2007; Nobile et al., 2010; Phua et al., 2012): i.e., 2500 grams and 30 points, respectively. For parental age at the child’s birth and parental education during the child’s first three years no firm empirical data or theoretical guidelines are available: therefore, we set arbitrary cut-off points at the 15th percentile of the distribution (i.e., 28 years old and 20 points, respectively). The adoption of these cut-offs led to 2 categories of risk, i.e., above (coded as ‘0’) and equal to/below (coded as ‘1’) the cut-off value, respectively. None of the above-defined variables had a missing values’ frequency > 10% and minor category’s frequency < 5% (data available upon request). References Abecasis, GR, Cardon, LR, Cookson, WO. (2000a) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66: 279-292. Abecasis, GR, Cookson, WO, Cardon, LR (2000b) Pedigree tests of transmission disequilibrium. Eur J Hum Genet 8: 545-551. DeCoster, J, Iselin, AMR, Gallucci, M. (2009) A conceptual and empirical examination of justification for dichotomization. Psychological Methods 14: 349-366. Fulker, DW, Cherny, SS, Sham, PC, Hewitt, JK. (1999) Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet 64: 259-267. Lasky-Su, J, Faraone, SV, Lange, C, Tsuang, MT, Doyle, AE, Smoller. JW, et al. (2007) A study of how socioeconomic status moderates the relationship between SNPs encompassing BDNF and ADHD symptom counts in ADHD families. Behav Genet 37: 487-497. Mascheretti, S, Bureau, A, Battaglia, M, Simone, D, Quadrelli, E, Croteau, J, Cellino, et al. (2013a) An assessment of gene-by-environment interactions in developmental dyslexia-related phenotypes. Genes Brain Behav 12, 47-55. Nobile, M, Rusconi, M, Bellina, M, Marino, C, Giorda, R, Carlet, O, et al. (2010) COMT Val158Met polymorphism and socioeconomic status interact to predict attention deficit/hyperactivity problems in children aged 10-14. Eur Child Adolesc Psychiatry 19: 549-557. Phua, DY., Rifkin-Graboi, A, Saw, SM, Meaney, MJ, Qiu, A. (2012) Executive functions of sixyear-old boys with normal birth weight and gestational age. PLoS One 7: e36502. Scerri, TS, Schulte-Korne, G. (2010) Genetics of developmental dyslexia. Eur Child Adolesc Psychiatry 19:179-197. Storey, JD. (2002) A direct approach to false discovery rates. J Royal Statistical Society, Series B 64: 479-498. Van den Oord. (1999) Method to detect genotype-environment interactions for quantitative trait loci in association studies. Am J Epidemiol 150: 1179-1187. van der Sluis, S, Dolan, CV, Neale, MC, Posthuma, D. (2008) A general test for gene-environment interaction in sib pair-based association analysis of quantitative traits. Behav Genet 38, 372-389. Zubrick, SR, Taylor, CL, Rice, ML, Slegers, DW. (2007) Late language at 24 months: an epidemiological study of prevalence, predictors, and covariates. J Speech Lang Hear Res 50: 15621592.