Exercise
advertisement

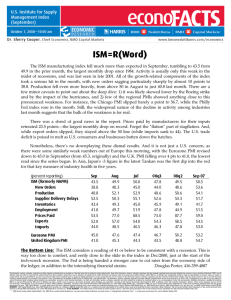
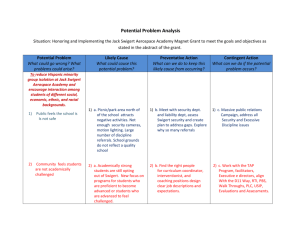
Exercise: Run Coupled Two-way Clustering using the CTWC Software (1,2) Biological data SKBR3 cells are derived of a breast tumor origin and express endogenously the H175R-p53 mutant. There are two close derived from this cell line: 1. Clone C8 - Conditional knockdown of mutant p53 protein in SKBR3. Addition of Doxycycline (a TET derivative) induces the relief of the repressor from pTERp53 RNAi that caused the induction p53 RNAi. Eventually a considerable knockdown in the expression of mutant p53 is observed. 2. Clone D11 - Express the Luciferase protein only was used as a control. Both clones were profiled before treatment (NT), and 48, 72 and 96 hours after addition of Doxycycline. For most conditions, there are three replicates – 2 technical repeats (1a and 1b) and one biological repeat (2). The table displays all chips. Names are in the format of chipname_cellline_time_batch_repeat, where celline is D8 or D11, time is NT/48/72/96, batch is c (for course) or s (for Shirly) and repeat is 1a, 1b (technical repeats) or 2 (biological repeat). D8 D11 NT 48hr 72hr D8_NT_s_1b D8_NT_c_1a D8_NT_c_2 D11_NT_s_2 D11_NT_c_1a D11_NT_c_1b D8_48h_s_1b D8_48h_c_1a D8_48h_c_2 D11_48h_c_1a D11_48h_c_1b D8_72h_s_1b D8_72h_c_1a 96hr D8_96h_s_1b D8_96h_c_1a D8_96h_c_2 D11_72h_s_2 D11_96h_c_1a D11_72h_c_1a D11_96h_c_1b D11_72h_c_1b Aim The aim of this exercise is to analyze the data using the Coupled Two-way Clustering (CTWC) software. Specifically, we would like to find a group of genes that allows us to cluster the samples by the cell line with good efficiency and purity. Instructions 1. All the data files are in the folder ‘C:/CTWC_SPIN/ex’. 2. View the file ‘var_500_rma.txt’ using Excel. This file contains the 500 genes with largest standard deviation across the samples (we chose to work with 500 genes in order to save time). 3. View the file ‘sample_labels.txt’ using Excel. This files contains the labels of the samples. The labels must be binary, ie 0 or 1. The first label C represents cell line 4. 5. 6. 7. (D8 = 0, D11 =1). The second label B represents batch (course = 0, before course = 1). The third label represents biological repeat. Question 1: What does the 1 in cell D3 in the labels file represent? Execute a CTWC analysis: a. Enter into the folder ‘C;/CTWC_SPIN/CTWC’and launch ‘run.bat’. b. A window will appear, press ‘try’. c. Maximize the CWC window. d. In the "Working Path" field set the working directory mentioned in (1). (You can use the browsing button on the right). e. In the "Data File Name" field browse (using the browsing button on the right) to the file mentioned in (2) and choose it. f. In the "Results Path" field choose the same path as for the "Working Path". (You can use the browsing button on the right). g. Since the data is already scaled, thresholded and log-transformed, the preprocessing panel can be skipped. h. In the "Labels Files" Samples field browse to the file sample_labels.txt. This will help us after the clustering (the labels are NOT used during the analysis) to find clusters that are correlated with known labels. i. Choose Depth 2 in the Samples "CTWC Depth" option. This will perform the coupled two-way analysis by clustering all the samples using each of the stable gene clusters that were found in the first iteration of CTWC. j. Finally... press "Start" and confirm. This executes the analysis. When the process will finish, you will be notified. You can follow the progress of the analysis at the command window or just wait a few minutes for the notification. Viewing the results of the analysis: a. Browse (using Windows browser) to your "Results Path", which was set in (6), and enter into the folder "Results-########". b. Open the file ‘index.html’. c. Question 2: How many stable clusters of genes were found when clustering all the 500 genes based on all the samples - G1(S1)? d. Question 3: How many stable clusters of samples were found when clustering all the samples based on all the genes - S1(G1)? e. Press the G1(S1) link to see the dendrogram and other figures related to clustering all the genes based on all the samples. f. Search for the gene TP53 in the G1(S1) page. g. Identify the stable clusters that contain this gene. The third column in the table lists the stable clusters that the gene belongs to. h. Identify the stable cluster(s) in the dendrogram and the corresponding blue box in the distance matrix. i. Question 4: What does the blue box along the diagonal mean? Find separation to D8/D11. a. Go back to the CTWC results main page (press "back" in the browser). b. Go to the "Clusters of Samples" section and identify the clustering operation S1(Gx), where Gx are the stable gene clusters you found in the previous section. c. For each of these: i. press the link ii. Identify stable samples clusters in the dendrogram. iii. Press in the cluster circle. A list of members should appear iv. Question 6: What is common to these members? v. Question 7: Is this separation more clear compared to S1(G1), look at the distance matrices ? d. Go back to the CTWC results main page and go to the last table: "Correspondence to External Labels of Samples". This table shows how the stable clusters of samples found in the unsupervised analysis relate to known labels. Look for the clusters of samples that you found above. e. Question 8: What is their purity and efficiency with respect to each cell line? CTWC server - http://ctwc.bioz.unibas.ch/ or http://ctwc.weizmann.ac.il/ References 1 2 3 Getz, G., Levine, E., and Domany, E. 2000. Coupled two-way clustering analysis of gene microarray data. PNAS 97:12079-12084. Getz, G. and Domany, E. 2003. Coupled two-way clustering server. Bioinformatics 19:1153-1154. Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R., and Korsmeyer, S. J. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. 30:41-47.