RT-PCR Master Mix Prep
advertisement

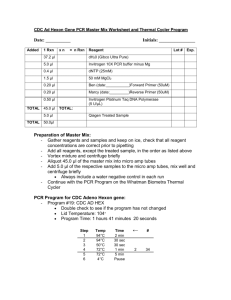
Human Rhinovirus (HRV) RT-PCR Two-Step PCR Set up Date: ________________ Initials: ________________ 1. Label 0.2ml thin-walled strip tubes with ID # and put on ICE. 2. Add 4 µl of random dodecamers to each tube. 3. Add 2 µl of 10 mM dNTP Mix 4. Add 18 µl of sample and/ or control into the appropriate tube. 5. Keep tubes on ice until they are placed into the thermocycler. 6. Run the samples under the following conditions: 65ºC 5 min. 4ºC ∞ 7. Keep samples on ice after removing from thermalcycler. 8. Make the Master Mix by combining the reagents listed below on ice. The reagent volume is multiplied by n = the number of samples + 1. Step 1 RT Reaction Mix Added TOTAL 1 Rxn 8.0 l 4.0 l 2.0 l 2.0 l Total Rxn 8.0 * n 4.0 * n 2.0 * n 2.0 * n 16.0ul 16.0 * n Reagent Lot # Exp. Invitrogen 5XM-MLV-RT Buffer DDT (0.1 M) RNasin (40U/ml) RNase Inhibitor Invitrogen M-MLV-reverse transcriptase (10U/ml) 9. Add 16.0 l of RT Rxn Mix into each tube containing sample and decamer. Mix by pipetting up and down. Change pipet tips between samples. 10. Quick spin tubes to remove air bubbles and put tubes into thermocycler. 11. Run under the following conditions. Step 1 2 3 Temperature Time 37°C 1 hr 70°C 5 min 4°C ∞ HRV Step Two PCR Date: ______________ Initials: ________________ Savolainen Primer (9565-reverse, 9895-forward) Protocol Reaction and cycling conditions as referenced in Mulders et al.,Molecular epidemiology of coxsackievirus B4 and disclosure of the correct VP1/2Apro cleavage site: evidence for high genomic diversity and long-term endemicity of distinct genotypes. J General Virology, 2000, 81:803-812. 1. Label 0.2ml thin-walled strip tubes with ID # and put on ICE. 2. Calculate the appropriate amount of the PCR master mix by calculating n (n=the number of reactions and controls plus one. Multiply the single Rxn volume by this number. PCR Master Mix Added TOTAL 1 Rxn xn = n Rxn Lot # 37.2 μl dH20 (Gibco Ultra Pure) 5.0 μl Invitrogen 10X PCR buffer minus Mg 0.4 μl dNTP (25mM) 1.5 μl 50 mM MgCl2 0.20 μl 9565-Forward Primer (50uM) 0.20 μl 9895-Reverse Primer (50uM) 0.50 μl Invitrogen Platinum Taq DNA Polymerase (5 U/µL) 45.0 μl Exp. TOTAL: 5.0 μl TOTAL Reagent First Strand cDNA 50.0μl 3. Assemble the PCR components on ice, and mix gently. 4. Aliquot 45ul of the PCR mix into the previously labeled 0.2ml strip tubes. 5. Using a multichannel pipet, pipet 5.0 l of the sample cDNA into the PCR mix and pipet up and down to mix. 6. Spin briefly and position tubes into the thermocycler. 7. Verify the program conditions before starting the program. Step 1 2 3 Temperature 94°C 94°C 60°C Time 5 min 15 sec 15 sec Return to Step# # Cycles 30 sec 2 40 (gradient 55-65) 4 6 72°C 4°C ∞ HRV Schnurr Primer (DK001, DK004) Protocol Reaction and cycling conditions as referenced in Kiang etl al., Assay for 5’ Noncoding Region Analysis of All Human Rhinovirus Prototype Strains, J Clin Microbiol, 2008, 46(11):3736-3745. 1. Label 0.2ml thin-walled strip tubes with ID # and put on ICE. 2. Calculate the appropriate amount of the PCR master mix by calculating n (n=the number of reactions and controls plus one. Multiply the single Rxn volume by this number. PCR Master Mix Added TOTAL 1 Rxn xn Reagent Lot # 37.2 μl dH20 (Gibco Ultra Pure) 5.0 μl Invitrogen 10X PCR buffer minus Mg 0.4 μl dNTP (25mM) 1.5 μl 50 mM MgCl2 0.20 μl DK001 Forward Primer (50uM) 0.20 μl DK004 Reverse Primer (50uM) 0.50 μl Invitrogen Platinum Taq DNA Polymerase (5 U/µL) 45.0 μl Exp. TOTAL: 5.0 μl TOTAL = n Rxn First Strand cDNA 50.0μl 3. Assemble the PCR components on ice, and mix gently. 4. Aliquot 45ul of the PCR mix into the previously labeled 0.2ml strip tubes. 5. Using a multichannel pipet, pipet 5.0 l of the sample cDNA into the PCR mix and pipet up and down to mix. 6. Spin briefly and position tubes into the thermocycler. 7. Verify the program conditions before starting the program. Step 1 2 3 4 6 Temperature 95°C 95°C 55°C 72°C 4°C Time 5 min 15 sec 15 sec 30 sec ∞ Return to Step# # Cycles 2 40