NMR Spectroscopy
advertisement

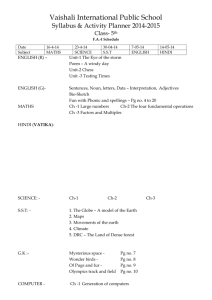
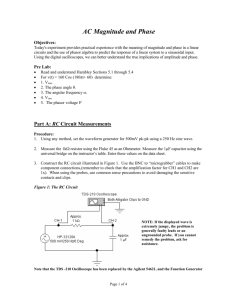
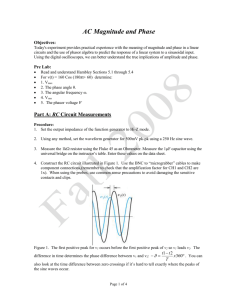
SUPPLEMENTARY MATERIAL Exploring Human Breast Milk composition by NMR-based Metabolomics Giulia Praticò1, Giorgio Capuani1, Alberta Tomassini1, Maria Elisabetta Baldassarre2, Maurizio Delfini1 and Alfredo Miccheli1* 1 Department of Chemistry, Sapienza University of Rome, Italy. Department of Gynecology, Obstetrics and Neonatology, Section of Neonatology and NICU, University of Bari, Italy 2 *Corresponding Author: Prof. Alfredo Miccheli, Department of Chemistry, Sapienza University of Rome, p.le Aldo Moro 5, 00185 Rome, Italy. Tel. ++39 06 49693270 Fax: ++39 06 49913214; e-mail: alfredo.miccheli@uniroma1.it Exploring Human Breast Milk composition by NMR-based Metabolomics Breast milk is a complex fluid evolutionarily adapted to satisfy the nutritional requirements of growing infants. In addition, milk biochemical and immunological components protect newborns against infective agents in the new environment. Human milk oligosaccharides (HMOs), the third most abundant component of breast milk, are believed to modulate the microbiota composition, thus influencing a wide range of physiological processes of the infant. Human milk also contains a number of other bioactive compounds the functional role of which has not yet been clearly elucidated. In this scenario, NMR-based metabolic profiling could provide a rapid characterization of breast milk composition thus allowing a better understanding of its nutritional properties. Keywords: NMR, 1H, HMO, milk metabolome, metabolic profiling Experimental NMR Spectroscopy The pulse sequence adopted for 1H NMR spectra acquisition was a presaturation pulse-single 90° detection pulse-acquire – relaxation delay. The relaxation delay was 7.5 s, while the presaturation pulse was applied for 2 s; the acquisition time needed to collect the 32 K points was about 5.5 s, therefore the proton nuclei different from the solvent ones were allowed to relax for 15 s, complying with the full relaxation condition after a 90 degree pulse. The length of the detection pulse was calibrated previously to the acquisition of each spectrum, spectral width was set to 5995.02 Hz (12 ppm), 64 scans were collected for each spectrum preceded by 4 dummy scans. Double quantum filter 1H-1H Correlation Spectroscopy (COSY) spectrum was recorded at 298 K with a spectral width of 15 ppm in both dimensions, employing a matrix of 4k x 400 data points, a repetition time of 2 s and 88 scans. Total Correlation Spectroscopy (TOCSY) experiment was performed at 298 K with a spectral width of 12 ppm in both dimensions, employing a matrix of 4k x 256 data points, a repetition time of 2 s, 112 scans. TOCSY spectra were acquired with several mixing times. In particular, they were optimized to evaluate the scalar correlation between H-1 and H-6 of the fucosylated moieties, on the basis of the long range scalar coupling constants. The 1H-13C Heteronuclear Single Quantum Correlation (HSQC) spectrum was recorded with the echo–antiecho phase-selective mode, with a spectral width of 12 and 200 ppm in proton and carbon dimensions respectively, employing a matrix of 4k x 400 data points for f2 and f1 dimensions, a repetition delay of 2 s and 96 scans. The average heteronuclear coupling constant was 145 Hz. The 1H-13C Heteronuclear Multiple Bond Correlation (HMBC) spectrum was obtained with 12 and 220 ppm spectral widths in proton and carbon dimensions respectively, employing a matrix of 4k x 400 data points for f2 and f1 dimensions, a repetition delay of 1.5 s and 96 scans. The long range heteronuclear coupling constant was 8 Hz, while the direct coupling constants comprised between 130 and 170 Hz were filtered. The Diffusion Order Spectroscopy DOSY experiment was acquired with a spectral width of 12 ppm for proton, 64 k data point, 144 scans, 4 dummy scans and a repetition delay of 5.16 s. The self-diffusion coefficients were evaluated with 32 gradient steps, a diffusion time (∆) of 350 ms and a gradient length (δ) of 2.5 ms.First paragraph style: use this for the first paragraph in a section. Table S1. List of metabolites identified in the hydro-alcoholic extract of human milk by 2D NMR TOCSY, HSQC and HMBC experiments. N° Name 1 2-oxoglutarate Group Fuc(α1-2) 2 2’FL Gal(β1-4) Fuc(α1-3)αGlc 3 3’FL Fuc(α1-3)βGlc Neu5Ac(α2-3) 4 3’SL 5 4-hydroxy-phenylacetate Neu5Ac(α2-6) Assignment 1H (ppm) β-CH2 γ-CH2 CH-1 CH-5 CH-4 CH3-6 CH-1 CH-4 CH-2 CH-1 CH-5 CH-3 CH-4 CH3-6 CH-1 CH-5 CH-3 CH-4 CH-2 CH3-6 CH-5 CH-6 CH-3 CH-3’ CH-2,6 ring CH-3,5 ring CH-5 CH-6 CH-3 CH-3’ 3.00 2.44 5.32 4.23 3.81 1.24 4.53 3.91 3.67 5.39 4.82 3.98 3.86 1.19 5.44 4.81 3.96 3.82 3.68 1.19 3.87 3.70 2.73 1.78 7.15 6.85 3.86 3.72 2.73 1.73 13C (ppm) 33.58 102.3 69.9 18.34 103.2 101.51 18.34 101.22 69.51 18.34 54.20 42.90 42.90 54.20 6 6’SL 7 Acetic Acid β-CH3 1.95 8 Alanine α-CH β-CH3 3.70 1.48 19.05 9 Carnitine N(CH3)3 3.23 56.91 10 Choline N(CH3)3 3.22 56.99 11 Citric Acid α,γ-CH α',γ'-CH CH-1 CH-4 CH-3 CH3-6 2.67 2.54 5.01 3.84 3.58 1.18 47.86 47.86 Fuc(α1-3)GlcNAc 12 Fucose moiety (1) 42.90 42.90 18.34 Fuc(α1-3)GlcNAc 13 14 15 16 17 18 19 20 21 22 CH-1 CH-5 Fucose moiety (2) CH-3 CH-4 CH3-6 Gal(β1-4)GlcNAc CH-1 CH-4 Galactose moiety (1) CH-6 CH-5 Gal(β1-4)GlcNAc CH-1 CH-4 Galactose moiety (2) CH-6 CH-5 Gal(β1-4)GlcNAc CH-1 CH-4 Galactose moiety (3) CH-6 CH-6’ CH-5 βGlc CH-1 CH-6 Glucose CH-6’ CH-3 CH-2 α-CH γ-CH2 Glutamic Acid β-CH β'-CH CH-2 ring Histidine CH-4 ring α-CH β-CH γ'-CH2 Isoleucine γ-CH2 β-CH3 δ-CH3 α-CH Lactic Acid β-CH3 Gal(β1-4) CH-1 CH-4 CH-6 CH-3 CH-2 αGlc CH-1 Lactosyl moiety CH-4 CH-6 CH-5 CH-3 CH-2 βGlc CH-1 CH-4 5.19 4.30 3.96 3.77 1.24 4.44 4.15 3.73 3.57 4.45 4.10 3.73 3.59 4.45 4.12 3.86 3.69 3.54 4.65 3.93 3.77 3.50 3.30 3.77 2.36 2.14 2.08 7.95 7.05 3.66 1.98 1.47 1.26 1.01 0.91 4.09 1.33 4.47 3.96 3.69 3.68 3.58 5.22 3.96 3.87 3.83 3.67 3.59 4.69 3.96 102.41 69.50 18.34 103.2 71.34 103.21 71.24 103.2 71.50 98.82 36.29 29.89 22.99 105.90 71.39 75.48 73.91 94.92 63.02 62.82 74.17 81.21 98.63 63.02 Fuc(α1-2) Fuc(α1-3)αGlc 23 LDFT Fuc(α1-3)βGlc Fuc(α1-2) Fuc(α1-4) 24 LNDFHI and branched GlcNAc(β1-6) Fuc(α1-4) Fuc(α1-3)αGlc 25 LNDFHII Fuc(α1-3)βGlc Gal(β1-4) 26 LNFPI and branched Fuc(α1-2) CH-6 CH-3 CH-2 CH-1 CH-5 CH-4 CH3-6 CH-1 CH-5 CH-3 CH-4 CH3-6 CH-1 CH-3 CH-4 CH3-6 CH-1 CH-5 CH-3 CH-4 CH3-6 CH-1 CH-5 CH-3 CH-4 CH-2 CH3-6 CH-1 CH-3 CH-6 CH-4 CH-5 CH3 CH-1 CH-3 CH-4 CH3-6 CH-1 CH-3 CH-4 CH-1 CH-3 CH-4 CH3-6 CH-1 CH-4 CH-2 CH-1 CH-3 CH3-6 3.81 3.68 3.30 5.29 4.26 3.81 1.28 5.40 4.82 4.00 3.83 1.19 5.46 3.98 3.83 1.23 5.16 4.35 4.01 3.77 1.29 5.03 4.88 3.98 3.82 3.65 1.26 4.61 4.14 3.83-3.96 3.75 3.54 2.08 5.03 3.96 3.82 1.18 5.38 3.98 3.79 5.43 3.98 3.79 1.14 4.51 3.91 3.54 5.32 4.01 1.24 63.16 77.27 76.69 102.30 18.34 101.50 18.34 101.21 18.34 102.70 69.51 18.34 106.23 25.30 100.81 18.34 101.71 101.50 18.34 105.81 GlcNAc CH3 2.06 Fuc(α1-4) 27 28 29 30 31 32 33 CH-1 5.03 CH-5 3.96 LNFPII CH-3 3.82 CH3-6 1.18 Fuc(α1-3)GlcNAc CH-1 5.11 CH-5 4.83 CH-3 3.91-4.01 CH-4 3.82 LNFPIII and branched CH3-6 1.19 Fuc(α1-3)GlcNAc CH-1 5.13 CH-5 4.85 CH-3 3.91 CH-4 3.82 CH3-6 1.19 Fuc(α1-3)αGlc CH-1 5.39 CH-5 4.82 CH-3 3.98 CH-4 3.86 CH3-6 1.19 LNPV Fuc(α1-3)βGlc CH-1 5.44 CH-5 4.81 CH-3 3.96 CH-4 3.82 CH-2 3.68 CH3-6 1.19 α-CH 3.68 ε-CH2 3.00 Lysine β-CH2 1.88 γ-CH2 1.69 δ-CH2 1.47 CH-2 ring 7.85 Me-Histidine CH-4 ring 7.06 N-CH3 3.09 CH-2 ring 7.80 Me-Histidine CH-4 ring 7.07 N-CH3 3.12 CH-1 4.06 CH-2,6 3.63 Myo-inositol CH-3,5 3.54 CH-4 3.28 100.81 18.34 102.40 69.50 18.34 101.51 18.34 101.51 18.34 101.22 69.51 18.34 42.29 29.06 75.12 34 N-acetylcarnitine N(CH3)3 3.21 56.80 35 N-Acetyl moieties (1) CH3 2.06-2.08 25.30 36 N-Acetyl moieties (2) CH3 2.03-2.05 25.04 37 Phenylalanine 38 Succinic Acid CH-3,4,5 ring 7.38-7.40 CH-2,6 ring 7.22 α-CH 3.92 β-CH 3.6 β’-CH 3.21 β-CH2 2.40 β-CH 4.26 α-CH 3.54 γ-CH3 1.34 CH-2,6 ring 7.20 40 Tyrosine CH-3,5 ring 6.90 CH-2,6 ring 7.19 41 U1 CH-3,5 ring 6.83 α-CH 2.19 40.40 γ-CH 1.57 42 Valeric Acid β-CH2 1.29 CH3 0.90 26.69 α-CH 3.59 β-CH 2.32 43 Valine γ-CH3 1.05 20.85 γ'-CH3 0.99 19.47 List of abbreviations: 2'FL, 2' fucosyllactose; Fuc Fucose; Gal Galactose; 3'FL, 3' fucosyllactose; Glc Glucose; 3’SL 3’sialyl-lactose; Neu5Ac N-acetylneuraminc acid; 6’SL 6’sialyl-lactose; LDFT, lactodifucotetraose; LNDFHI- II, lacto-N-difucohesaose I-II, LNFPI –III, V, lacto-N-fucopentaose I-III, V; GlcNAc N-acetylglucosamine. 39 Threonine Table S2. Quantification of shared metabolites in milk group 1 (Se+/Le+) and milk group 2 (Se-/Le+) . Milk Group 1 (Se+/Le+) (µM) Isoleucine Valine Fuc (α1-3)+Fuc(α1-4) Lactic Acid Alanine + Lysine Acetic Acid GlcNAc moiety 1 GlcNAc moiety 2 GlcNAc moiety 3 GlcNAc moiety 4 Glutamic Acid Succinic Acid Citric Acid N-acetylcarnitine Choline Carnitine Myoinositol Lactose Fucose moiety 1 Fuc(α1-4) LNFPIII Fucose moiety 2 Phenylalanine 3’FL LNDFH II GlcNAc total 0.8 14.8 2027.1 72.1 139.3 52.5 1384.5 1663.8 1261.8 1575.0 1045.6 20.9 1418.8 108.7 414.8 362.8 1525.9 143800.5 339.0 1260.1 534.7 1430.4 1387.7 1051.6 53.2 8974.8 Milk Group 2 (Se-/Le+) (µM) (1.2) (18.8) (1057.5) (50.5) (68.6) (37.0) (963.1) (755.5) (606.8) (854.5) (416.3) (20.1) (697.5) (61.8) (274.2) (116.9) (585.2) (23088.2) (82.3) (451.6) (266.9) 0.8 18.6 5875.9 96.5 132.1 56.6 2897.6 1948.3 2162.8 1436.0 966.7 19.4 1633.8 75.2 573.6 364.6 2125.1 147288.8 426.2 1756.0 543.4 (80.8) (6.6) (2632.4)** (70.9) (29.4) (17.5) (1139.6)* (1213.2) (1093.1) (1020.1) (161.4) (19.7) (1049.3) (46.9) (202.6) (171.0) (863.6) (57686.1) (139.8) (907.2) (328.7) (309.6) (1224.9) (582.4) (40.1) (3064.2) 1163.5 783.9 2571.7 365.5 10293.1 (748.1) (1151.2) (1031.7)* (249.3)* (3707.3) Data are presented as medians of concentration (µM) and ranges from the 1st to the 3rd quartile. *p<0.001; ** p<0.01 between values. List of abbreviations: Fuc Fucose; 3'FL, 3' fucosyllactose; LNDFH II, lacto-N-difucohesaose II, LNFPIII, lacto-Nfucopentaose III; GlcNAc N-acetylglucosamine. Figure S1. 1H-1H TOCSY NMR experiment of the polar extract of mature breast milk; the figure displays the TOCSY spectral pattern of fucose units of 2’fucosyllactose (2’FL) and 3’fucosyllactose (3’FL), whose resonances correspond exactly with those of lacto-N-fucopentaose V (LNFPV), due to the strong structural similarity. List of abbreviations: 2'FL, 2' fucosyllactose; 3'FL, 3' fucosyllactose; LDFT, lactodifucotetraose; LNDFHII, lacto-N-difucohesaose II, LNFPI, lacto-Nfucopentaose I. Figure S2. 1H-13C HSQC NMR experiment of the polar extract of mature breast milk, detail of H-1 fucose resonances. List of abbreviations: 2'FL, 2' fucosyllactose; 3'FL, 3' fucosyllactose; LNDFHI, lactoN-difucohesaose I, LNFPIII, lacto-N-fucopentaose III.