Nitrogen Acquisition and Amino Acid Metabolism
advertisement

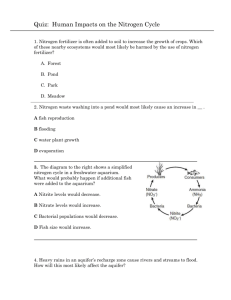
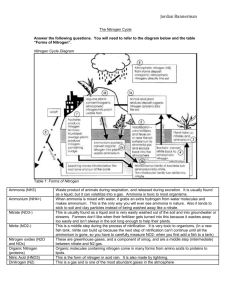
Fundamentals I: 11:00-12:00 Monday, August 30, 2010 Dr. Larry Delucas I. Nitrogen Acquisition and Amino Acid Metabolism Scribe: Karin Tran Proof: Peyton Yarbrough Page 1 of 5 Nitrogen Acquisition and Amino Acid Metabolism [S1] a. Nitrogen is a critical component of amino acids, nucleotides, and proteins. b. How do we get nitrogen? c. Where does nitrogen go in the environment? d. How is it put in a form that we, as eukaryotes, can use it? e. Not going to test on reaction mechanisms. Just know the general process of how some of the amino acids are made and how we utilize nitrogen and how nitrogen is obtained from the environment. II. Key Concepts/Amino Acid Metabolism [S2] III. Nitrogen Fixation and Assimilation [S3] IV. Essential Questions [S4] a. Understand the general concepts, not memorization b. Essential questions are highlighted in red. V. Outline [S5] a. He read the questions off the slide VI. How Do Various Organic Forms of Nitrogen Arise? [S6] a. Amino acids and nucleotides are critical for biological function. b. We (eukaryotes) can’t take inorganic forms of nitrogen and put it in the reduced form we need to make amino acids. c. Prevalent forms of nitrogen i. atomospheric nitrogen (78% of atmosphere) ii. nitrate anions (NO3-) iii. These are oxidized forms of nitrogen. iv. We have to have reduced form. d. There are 2 ways to convert the oxidized forms into reduced states. i. Nitrogen Fixation – deals with gaseous nitrogen (N2) ii. Nitrate Assimilation - deals with anions NO3- and NO2iii. These processes are done by plants and microorganisms, NOT us. iv. In both cases, ammonium ions are formed which we can use to create amino acids VII. Which Metabolic Pathways Allow Organisms to Live on Inorganic Forms of Nitrogen? [S7] a. Nitrogen is always being cycled. b. We excrete nitrogen in a reduced form (ie. urea) which gets cycled back into an oxidized form by certain bacteria. The oxidized form is then cycled back into a different reduced form for us to utilize once again. c. The process of reducing nitrate to an ammonium ion is called nitrate assimilation. i. This process is done by plants, fungi, and certain bacteria. ii. It’s a 2-step pathway that involves 2 enzymes d. Nitrate assimilation is responsible for 99%. Nitrogen fixation is responsible for 1%. VIII. Major Pathways for N Acquisition [S8] a. Through assimilation, inorganic forms of nitrogen like nitrate (NO 3-) gets reduced to nitrite (NO2-). b. The fully reduced form is ammonium ion with protons on it (NH 4+). c. Through a series of reactions, NH4+ will get cycled into organic nitrogen that’s combined with other atoms. d. Through fixation, gaseous nitrogen can be reduced to NH4+ ions which also get cycled into organic nitrogen. e. Nitrifying bacteria (autotrophs) can convert NH4+ back to a more oxidized state through the nitrification process. f. Denitrifying bacteria can create gaseous nitrogen through a process called denitrification. i. This process is important because there was concern over the uncontrolled plant growth in lakes and ponds due to the buildup of oxidized forms of nitrogen. (acts like a fertilizer) ii. These bacteria play a role by shifting those oxidized forms back to gaseous nitrogen. g. Nitrogen fixation can also occur through lightning strikes (~1%) IX. Nitrate Assimilation is the Principal Pathway for Ammonium Biosynthesis [S9] a. There are 2 enzymes involved in nitrate assimilation. i. Nitrate reductase takes nitrate with 2 H atoms and 2 e- and converts it to nitrite and H2O is given off. Fundamentals I: 11:00-12:00 Scribe: Karin Tran Monday, August 30, 2010 Proof: Peyton Yarbrough Dr. Larry Delucas Nitrogen Acquisition and Amino Acid Metabolism Page 2 of 5 ii. Nitrite reductase requires 6 e- and 8 H atoms to reduce NO2- to NH4+. H2O is given off. X. Nitrate Assimilation [S10] a. In the protein complex of nitrate reductase, there are a series of e- accepting components (FADH, Heme, Mo-Pterin cofactor) b. The complex transfers e- from NADH and ends up in the nitrite form c. Describing enzyme based on figure (a) at the bottom left i. Yellow and blue area: parallel beta sheets surrounded by alpha helices illustrate an alpha/beta protein ii. Bottom right blue area: anti-parallel beta sheets in the form of a beta barrel iii. Bottom left red: anti-parallel beta sheets with alpha helices on top and below (green) d. Being able to pick these themes out from within the structure is useful because a pocket is usually formed for something to sit and bind very closely with atoms around it (ie. Molybdenum) XI. Nitrate a. b. c. d. Assimilation is the Principal Pathway for Ammonium Biosynthesis [S11] This is just a prosthetic group but there is a protein that surrounds it and is in close association with it. The Molybdenum cofactor is unique cofactor found in many enzymes. It is surround by a shell of several amino acids that interact with it. It’s able to accept electrons just like the other components of the enzyme complex i. (refer back to previous slide) ii. The e- are passed along the enzyme complex from component to component. iii. The Fe and other metals can coordinate with the negative charge as the e- are passed along. iv. The nitrate binds into the pocket to the right of Molybdenum and makes an interaction with that metal. XII. Nitrate Reductase [S12] a. Nitrate reductase is a homodimer. b. Electrons pass from NADH portion toward the Molybdenum site. c. There are hinge regions that move as electrons pass. d. This movement allows for the accommodation of the electrons being close enough to carry over to the next region. e. Electrons are passed into the region where nitrate is bound by movements of the protein. f. There are 2 different hinge regions. XIII. The Reaction of Nitrite Reductase [S13] a. Nitrite reductase in plants is usually activated by light. b. Ferredoxin can get easily reduced. i. It is an Fe atom that has substituents around it that allow it to easily accommodate electrons. c. Electrons from ferredoxin are transported to a cluster of 4 Fe and 4 S which links to the siroheme transport center d. At this point, nitrite gets reduced fully to ammonium e. Structure of siroheme i. Fe can coordinate directly with nitrite ii. The carboxyl groups on the outside can donate H atoms so there is a passing of double bonds from one component to another iii. Protons are released and end up hooking onto the nitrogen when it acquires 2 electrons. iv. Build hydrogens around nitrogen to make ammonium ion. f. Bottom line: You have a transfer of electrons from groups that can accept electrons through the nitrite reductase enzyme to where the nitrite ion is bound to create ammonium. g. The prosthetic group siroheme plays critical role in which it can also donate protons. XIV. Nitrite Reductase [S14] a. Example of nitrite reductase with iron-sulfur cluster and siroheme b. It is an alpha/beta structure XV. Nitrate Assimilation is the Principal Pathway for Ammonium Biosynthesis [S15] a. Plant and fungal nitrate reductases are about 200 kD and are homodimers. i. On the N-terminus, the Mo cofactor is the first component made ii. There’s a hinge region, cytochrome b group, FAD group iii. The process starts on the end with the NAD(P)H. b. NitrIte reductase is a much smaller molecule. Fundamentals I: 11:00-12:00 Scribe: Karin Tran Monday, August 30, 2010 Proof: Peyton Yarbrough Dr. Larry Delucas Nitrogen Acquisition and Amino Acid Metabolism Page 3 of 5 i. Plant nitrite reductase 1. There is an electron donor. 2. Nitrite is bound to the siroheme group until it becomes the nitronium ion. ii. Fungal nitrite reductase looks like the nitrate reductase of plants and fungi with an FAD and NAD(P)H group iii. There is also a cysteine rich region and a FeS-siroheme group. c. This slide just shows the organization of the sequences of each enzyme. XVI. Organisms Gain Access to Atmospheric N2 Via the Pathway of Nitrogen Fixation [S16] a. Nitrogen fixing bacteria can be free living or symbiotic with eukaryotic organisms. b. Bacteria like Rhizobia hook to leguminous plants and help each other i. It can provide a more reduced form of nitrogen for the plant, and the plant is helping the bacteria by giving it nitrogen bound to other molecules to use as metabolites. ii. The bacteria has start the process of nitrogen fixation in order for this relationship to occur. c. 4 key components are required i. All nitrogen fixing bacteria utilize nitrogenase enzyme. ii. A reductant, usually reduced ferredoxin iii. Energy (ATP) in order to make process occur iv. Has to be anaerobic because nitrogen becomes an acceptor for e- and the presence of oxygen would inhibit that. v. There are also regulatory controls. 1. ADP inhibits 2. High concentration of ammonium ions inhibits expression. XVII. Figure 1. Global Nitrogen Cycle [S17] a. Use this chart to quiz yourself. XVIII. The Nitrogenase Reaction [S18] a. There are 2 components i. Nitrogenase Reductase ii. Nitrogenase b. The whole complex is call the nitrogenase complex but there are 2 enzyme components c. The process takes gaseous nitrogen with 10 H and 8 e- and creates 2 molecules of ammonium ions, and gaseous H is released. d. This reaction takes a lot of energy (16 ATP) XIX. Nitrogenase Complex [S19] a. The nitrogenase complex is a homodimer. b. There is nitrogenase reductase (blue) on either end of the complex c. The green is the nitrogenase. d. FeS cluster = yellow, ADP = orange, FeMo = cyan, P cluster = red XX. Organisms Gain Access to Atmospheric N2 Via the Pathway of Nitrogen Fixation [S20] a. Why does the reaction take so much energy? i. Nitrogen has a strong triple bond that must be broken. ii. One way to break the bond is with lightning, but this occurs only 1% of the time iii. The main way is to use ATP for energy. b. The reaction is favored but you need energy to get passed the transition state. c. For each e- being transferred, you need 2 ATP. d. Since 8 e- are being transferred, 16 ATP is needed. XXI. The Metal Clusters of Nitrogenase [S21] a. There are 2 metal clusters of nitrogenase complex. b. The bottom one has Molybdenum which has 7 Fe and 9 S (FE7S9). One Fe is shared with another molecule. c. The upper one has 8 Fe complexed with 7 S (FE8S7). XXII. The Nitrogenase Reaction [S22] a. ATP hydrolysis has to occur to give energy for the reaction. b. The first process is transferring e- to the P cluster from the nitrogenase reductase enzyme that interacts with nitrogenase. Fundamentals I: 11:00-12:00 Scribe: Karin Tran Monday, August 30, 2010 Proof: Peyton Yarbrough Dr. Larry Delucas Nitrogen Acquisition and Amino Acid Metabolism Page 4 of 5 i. ATP creates energy to cause e- to go from the nitrogenase reductase over to the P cluster. ii. ATP is converted to ADP. iii. This causes a conformational change in the nitrogenase reductase so it can no longer bind to nitrogenase. iv. The enzyme diffuses away and allows another nitrogenous reductase with ATP to bind and contribute its ec. The P cluster transfers the e- ultimately to the FeMo part of the nitrogenase enzyme. d. Build up of e- occurs at the FeMo where there is also the triple bonded nitrogen interacting with the Fe. e. Nothing happens until there is enough e-. No intermediates are formed. f. 3 molecules of ammonium are created per second (We need way more.) g. The nitrogenase enzyme is extremely slow, so one way to compensate is to have a multitude of nitrogenase enzyme. i. For organisms that have this complex, 5% of the their genome is making the nitrogenase enzyme (huge percentage) h. The transfer of e- by nitrogenase reductase to the P cluster becomes the slow part of the reaction. This limits the rate of the whole reaction. i. Once 8 e- are built up in the FeMo region, the ammonium ion can be made directly from gaseous nitrogen. XXIII. The Structure of Nitrogenase [S23] XXIV. The Structure of Nitrogenase (Gray) [S24] a. Not going to ask question about structure. b. Slides are added to see the close association of the different substituents. c. They are within H bond distance, 2.8 angstroms apart so e- can easily be shared and shifted from one substituent to another. XXV. The Structure of Nitrogenase (Gray) [S25] a. Even better slide to show the distances b. There’s a lot of conformational changes. c. Nitrogenase reductase is similar to G proteins. d. Because of these huge conformational changes, it can no longer interact with nitrogenase, so it diffuses away. XXVI. Regulation of Nitrogen Fixation [S26] a. It can be regulated through gene expression. b. There is direct feedback inhibition of the nitronium ions on the genes that express the nitrogenase molecule. c. There is feedback inhibition of ADP. High amounts of ADP means there is a high amount of ammonium ions, so feedback inhibition occurs. d. A conversion from an active state to a nonactive state can occur through direct covalent bond interaction. i. ADP ribose unit binds to the nitrogenase enzyme are Arg101 and inactivates it. ii. A conformational change occurs to inactivate the enzyme. XXVII. What is the Metabolic Fate of Ammonium? [S27] a. We want to be able to take the ammonium ions and make amino acids or nucleotides. b. Only 3 enzymes are responsible i. Glutamate dehydrogenase ii. Glutamine Synthetase iii. Carbamoyl-phosphate synthetase 1 1. A mitochondrial enzyme 2. One of the first steps that leads a certain metabolite into the urea cycle iv. The first 2 enzymes are responsible for a major percentage of ammonium ions getting converted into different substituents that can lead into amino acids. XXVIII. Glutamate Dehydrogenase [S28] a. Alpha-ketoglutarate is converted to glutamate via reductive amidation. b. An NH3+ group replaces the C=O. c. Energy is provided by NADPH. XXIX. Glutamine Synthetase [S29] a. There are 2 parts i. Energy is provided by ATP Fundamentals I: 11:00-12:00 Scribe: Karin Tran Monday, August 30, 2010 Proof: Peyton Yarbrough Dr. Larry Delucas Nitrogen Acquisition and Amino Acid Metabolism Page 5 of 5 1. When ATP interacts with glutamate, it phosphorylates the COOH group ii. Ammonium binds and causes the phosphate to leave. b. The result is glutamine, one of our amino acids. c. Glutamine is a precursor to other amino acids that we can make (nonessential amino acids). XXX. The Carbamoyl-Phosphate Synthetase [S30] a. This reaction is an early step in the urea cycle. b. Bicarbonate is needed. c. Bicarbonate is used to combine with ammonium ion to get the products of the reaction (see slide). d. This enzyme is a synthetase enzyme – ATP is used for energy e. Synthase enzymes perform synthesis reactions but do not use ATP. XXXI. What Regulatory Mechanisms Act on E. coli Glutamine Synthetase? [S31] a. In E. coli, there is an example about how Glutamine Synthetase works. b. Glutamine synthetase is huge (600 kD). c. It has 6 peptides on top and 6 below in a hexagon ring. They are 2 hexagons stacked on top of each other. d. It is regulated 3 ways i. Feedback inhibition ii. Covalent modification iii. Regulation of gene expression and protein synthesis XXXII. The Subunit Organization of Bacterial Glutamine Synthetase [S32] a. There are 2 different views of the enzyme shown. b. On the top view, the active component is between the red and yellow portions. c. You need 2 of the monomers forming the dodecohexamer for the enzyme to work. d. A monomer by itself has no activity. e. Through gene duplication E. coli arrived at the dodecohexamer to quickly do its job rather than just having a dimer. XXXIII. Allosteric Regulation of Glutamine Synthetase [S33] a. Its process is to make glutamine but it’s regulated a number of ways. b. 9 amino acids serve as feedback inhibitors. c. They would inhibit the process of this enzyme creating glutamine. d. Other substituents (ie. CTP) that come off as glutamine does its job also do feedback inhibition. e. Amino acids made from glutamine acts as feedback inhibitors on the synthesis of glutamine. XXXIV. Glutamine Synthetase if Regulated by Covalent Modification [S34] a. Technical name: ATP Glutamine Synthetase Adenyly Transferase b. Adenylyl transferase transfers adenylyl group onto glutamine synthetase and inactivates it. c. This happens at Tyr397 for each of the monomers. d. The Tyr gets modified which inactivates the enzyme. e. There are 12 glutamine synthetase monomers so it can be inactivated 12 times. f. The more the enzyme is adenlylated, the more it is inhibited. XXXV. The Cyclic Cascade System Regulating the Covalent Modification of GS [S35] a. There are 2 proteins: P2A complex and P2D complex b. The P2D complex bound to adenylyl transferase functions to de-adenylate. i. When it binds to glutamine synthetase, the reverse reaction occurs. ii. De-adenlyation makes the enzyme more active. c. The P2A complex makes the reaction go to the right and adenylation occurs. (Refer to previous slide.) d. Alpha-ketogluarate and glutamine also regulate the process. i. High levels of alpha-ketoglutarate will cause de-adenylation to occur. ii. High levels of glutamine will cause adenylation to inactivate the enzyme. XXXVI. Glutamine Synthetase is Regulated Through Gene Expression a. We don’t have this slide. b. He won’t ask a question on this. [End 50:00 mins]