supplemental materials and methods
advertisement

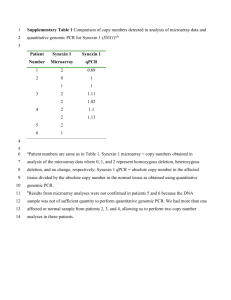
SUPPLEMENTAL MATERIALS AND METHODS Identification of SNPs physically linked to previously identified genetic markers: Previous mapping efforts of the allorecognition trait relied on two Amplified Fragment Length Polymorphisms (AFLPs), designated 194 and 174, one co-dominant PCR marker designated 18, and one dominant PCR marker, R28, that together defined a 3.4 cM region. A fifth dominant AFLP marker (F29) was also identified but was determined to segregate only with the f haplotype and therefore could not be used for mapping in the backcross since all individuals carried the f haplotype (CADAVID et al. 2004). Poudyal et al. (2007) described the cloning of the AFLP 194 marker and extension of this genomic sequence in the r haplotype as well as the identification of the sequence of a corresponding f allele. Analysis of that sequence revealed only one SNP. Therefore additional primers were designed to extend this sequence by the Universal GenomeWalker™ method (Clontech Laboratories). A 2.2 kb extended sequence of this region identified an additional 12 SNPs in the 194 genomic region. In the marker 18 sequence, three SNPs were identified that were suitable for the hME assay. No SNPs were known for the short (80bp) r-segregating sequence of marker 28. The GenomeWalker™ method was used to extend the sequence in this region and genomic products were sequenced and PCR primers were designed to amplify a 0.8 kb region in ff/ff and rr/rr DNA. Sequencing of these products identified a total of eight SNPs. Marker 174 was a 247 bp AFLP present only in the r haplotype. In order to clone and sequence 174, the fragment was generated as described by Cadavid et al. (2004) with the exception that the primers used in the selective amplification contained no fluorescent label. The AFLP fragments were separated by gel electrophoresis using 3% Agarose, Resolute GPG (American Bioanalytical, Natick, MA). Those fragments corresponding to the predicted size of the 174 marker (247 bp) were excised from a gel and purified using the Qiaquick® Gel Extraction Kit (Qiagen, Valencia, CA) and cloned into the pCRIITOPO TA vector (Invitrogen) and transformed into Subcloning Efficiency™ DH5™ Competent Cells (Invitrogen). Because these clones still represented independent AFLP products of similar size, the clones containing 174 were identified by performing a PCR using the fluorescence-labeled primers originally used for selective amplification in the AFLP procedure (CADAVID et al. 2004). The products were run on a 6% acrylamide gel and analyzed to identify clones containing a 247 bp fragment, which were then sequenced. The insert of a positive clone was also used as a probe in a Southern blot containing selective amplification products (from the AFLP procedure) of 3 homozygous ff/ff individuals and 4 heterozygous rr/ff individuals to verify the correct band had been identified. Primers designed to amplify the 174 r sequence also amplified a fragment in ff/ff DNA. This product was subsequently used to physically map and clone the alr2 chromosomal region using fosmids and BAC libraries described in NICOTRA (2007). End-sequences generated in the course of this study were used to design primers that amplified corresponding fragments from the f and r haplotypes. These PCR products were sequenced and a total of 25 SNPs were identified. Marker 29 was a dominant AFLP marker linked to the f haplotype that was cloned and sequenced by Cadavid et al. (2004). This DNA was used as a probe to screen the fosmid library described by Nicotra (2007). A fosmid (pYU1527) was recovered and end-sequenced to design probes that were hybridized to the ff/ff genomic BAC library. Thirteen hybridizing BACs were identified and end-sequenced. PCR primers were designed to the end sequence and tested on rr/rr genomic DNA. These products were sequenced and compared to the ff/ff sequence, which led to the identification of 36 SNPs. LITERATURE CITED CADAVID, L. F., A. E. POWELL, M. L. NICOTRA, M. MORENO and L. W. BUSS, 2004 An invertebrate histocompatibility complex. Genetics 167: 357-365. NICOTRA, M. L., 2007 Positional cloning of a cnidarian allorecognition locus. Ph.D. Thesis, Yale University, New Haven, CT. POUDYAL, M., S. ROSA, A. E. POWELL, M. MORENO, S. L. DELLAPORTA et al., 2007 Embryonic chimerism does not induce tolerance in an invertebrate model organism. Proc. Natl. Acad. Sci. USA 104: 4559-4564.