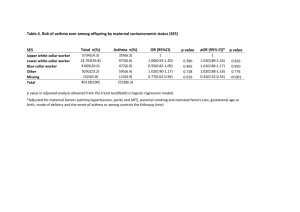
cea4078-sup-0001-TableS1
Gene
PTGS1
PTGS2
ALOX5AP
ALOX5
ALOX12
ALOX15
LTC4S
PTGDR
PTGER1
PTGER2
PTGER3
PTGER4
Chromosome
9
1
13
10
17
17
5
14
19
14
1
5
SNP rs# rs5789 rs1236913 rs689466
Position
844C>A (Leu237Met)
157C>T (Trp8Arg)
-1329T>C
-297C>G
Other names
Ex7+31C>A, c.709C>A c.22C>T
-1195A>G,
A496G
-296C>G rs5270 rs1132340 218A>G
-1700G>A rs4986832 -1699A>G rs6413416 (*) rs1126667
-1753G>A
782 A>G (Gln261Arg)
-1752A>G
Ex6-26A>G rs434473 rs2255888
965A>G (Asn322Ser)
-635C>T rs7220870 rs3892408 (*)
-272C>A
729C>T (Val239Met)
Ex8+14A>G
-634G>A
-271T>G
A-444C rs730012 rs34236606 rs8004654 rs803010
-444A>C
-613 T>C
-549 T>C
-441 T>C rs11157907 rs3810253 rs3810255 rs1254598 rs1353411
-197 T>C
-1013 A>C
-804 C>T
-72 G>A
-166 G>A rs2075797 rs1254600 rs7551789 rs13186505 rs4133101 rs4957341
-616 C>G
-814 A>G
-1709 A>T
-936 G>A (*)
-1057 T>C
40668049 G>A (*)
MAF
0.07
0.32
0.08
0.47
0.28
0.43
0.25
0.24
0.40
0.13
0.22
0.16
0.22
0.18
0.06
0.17
0.02
0.09
0.21
0.19
0.44
0.13
0.16
0.07
0.28
0.36
0.10
Previous studies
[2]
[3,4]
[6]
[7]
[10-19]
[23-24]
[23-26]
[23-24]
[23-24]
[26]
[26]
[27]
[27]
[27]
[27]
[26]
[8]
[8]
[8]
[8]
[26]
Functional evidences
[1]
[1]
[5]
[5]
Predicted functional effects
Missense (conservative); splicing regulation.
Missense (conservative); splicing regulation.
Promotor/regulatory region: changes in
TFBS.
Promotor/regulatory region: changes in
TFBS.
MicroRNA binding site.
Promotor/regulatory region: changes in
TFBS.
Upstream with no known function.
TFBS.
Splicing regulation.
[20-22]
[23]
[23]
[23]
[24,25]
Splicing regulation.
Upstream with no known function.
TFBS.
Promotor/regulatory region: changes in
TFBS.
Missense (conservative); splicing regulation.
Upstream with no known function.
TFBS.
TFBS
Promotor/regulatory region: changes in
TFBS.
Promotor/regulatory region: changes in
TFBS.
TFBS
TFBS
TFBS
TFBS
Promotor/regulatory region: changes in
TFBS.
Promotor/regulatory region: changes in
TFBS.
TFBS
TFBS
TFBS
TFBS
TFBS
PTGFR
CYSLTR1
1
X rs3753380 rs320995
-562 T>C
927 C>T (Phe309Phe)
0.33
0.24
[26]
[28-31] [21]
TFBS
Sense/synonymous; splicing regulation.
Downstream with no known function.
CYSLTR2 13 rs912277 g49283651 0.07
[7] rs912278 2534 A>G 0.42
[7]
Splicing regulation; microRNA binding site
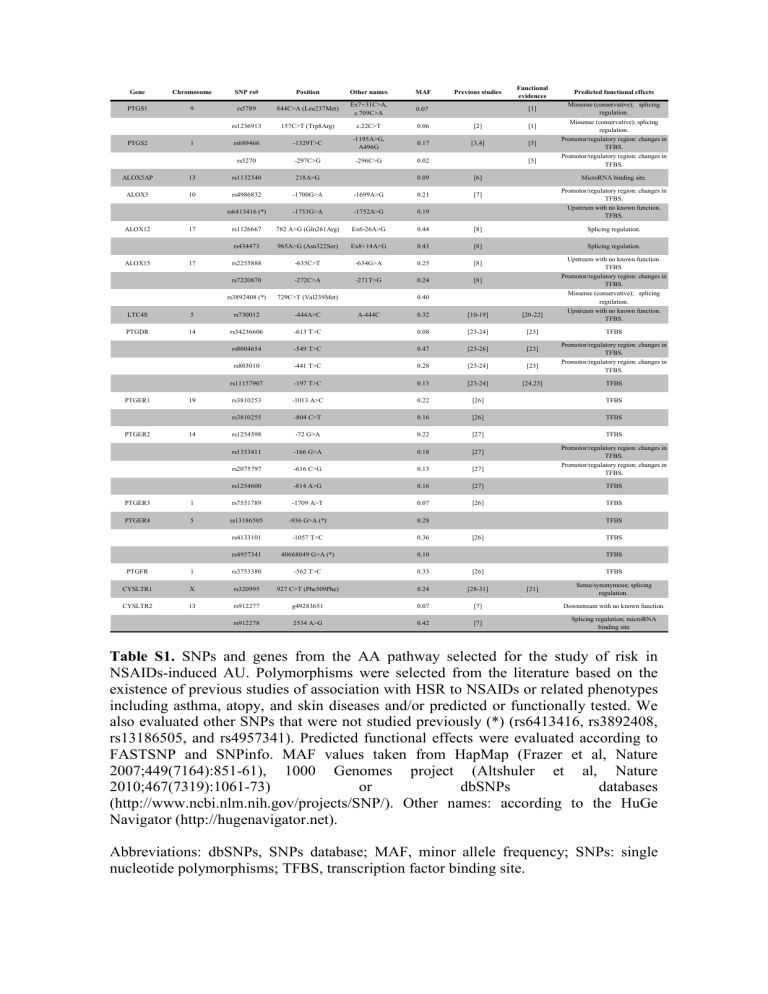
Table S1. SNPs and genes from the AA pathway selected for the study of risk in
NSAIDs-induced AU.
Polymorphisms were selected from the literature based on the existence of previous studies of association with HSR to NSAIDs or related phenotypes including asthma, atopy, and skin diseases and/or predicted or functionally tested. We also evaluated other SNPs that were not studied previously (*) (rs6413416, rs3892408, rs13186505, and rs4957341). Predicted functional effects were evaluated according to
FASTSNP and SNPinfo. MAF values taken from HapMap (Frazer et al, Nature
2007;449(7164):851-61), 1000 Genomes project (Altshuler et al, Nature
2010;467(7319):1061-73) or dbSNPs databases
(http://www.ncbi.nlm.nih.gov/projects/SNP/). Other names: according to the HuGe
Navigator (http://hugenavigator.net).
Abbreviations: dbSNPs, SNPs database; MAF, minor allele frequency; SNPs: single nucleotide polymorphisms; TFBS, transcription factor binding site.
SUPPLEMENTARY REFERENCES:
[1]Lee CR, Bottone FG Jr, Krahn JM, Li L, Mohrenweiser HW, Cook ME, et al.
Identification and functional characterization of polymorphisms in human cyclooxygenase-1 (PTGS1). Pharmacogenet Genomics 2007;17:145-60.
[2] Shi J, Misso NL, Duffy DL, Bradley B, Beard R, Thompson PJ, et al.
Cyclooxygenase-1 gene polymorphisms in patients with different asthma phenotypes and atopy. Eur Respir J 2005;26:249-56.
[3] Zhang X, Miao X, Tan W, Ning B, Liu Z, Hong Y, et al. Identification of functional genetic variants in cyclooxygenase-2 and their association with risk of esophageal cancer. Gastroenterology 2005;129:565-76.
[4] Dong J, Dai J, Zhang M, Hu Z, Shen H. Potentially functional COX-2-1195G>A polymorphism increases the risk of digestive system cancers: a meta-analysis. J
Gastroenterol Hepatol 2010;25:1042-50.
[5] Skarke C, Schuss P, Kirchhof A, Doehring A, Geisslinger G, Lötsch J.
Pyrosequencing of polymorphisms in the COX-2 gene (PTGS2) with reported clinical relevance. Pharmacogenomics 2007;8:1643-60.
[6] Kim SH, Choi JH, Holloway JW, et al. Leukotriene-related gene polymorphisms in patients with ASA-induced urticaria and ASA-intolerant asthma: differing contributions of ALOX5 polymorphism in Korean population. J Korean Med Sci 2005; 20:926–31.
[7] Klotsman M, York TP, Pillai SG, Vargas-Irwin C, Sharma SS, van den Oord EJ, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics 2007;17(3):189-96.
[8] Lee HY, Kim SH, Ye YM, Choi GS, Park HS. Lack of association of ALOX12 and
ALOX15 polymorphisms with aspirin-exacerbated respiratory disease in Korean patients. Ann Allergy Asthma Immunol 2009;103:84-6.
[9] Sanak M, Simon HU, Szczeklik A. Leukotriene C4 synthase promoter polymorphism and risk of aspirin-induced asthma. Lancet 1997;350:1599-600.
[10] Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, Higashi N, et al.
Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirininduced asthma. J Allergy
[11] Choi JH, Park HS, Oh HB, Lee JH, Suh YJ, Park CS, et al. Leukotriene-related gene polymorphisms in ASA intolerant asthma: an association with a haplotype of 5lipoxygenase. Hum Genet 2004;114:337-44.
[12] Kim SH, Yang EM, Park HJ, Ye YM, Lee HY, Park HS. Differential contribution of the CyslTR1 in patients with aspirin hypersensitivity. J Clin Immunol 2007;27:613-9.
[13] Mastalerz L, Nizankowska E, Sanak M, Mejza F, Pierzchalska M, Bazan-Socha S, et al. Clinical and genetic features underlying the response of patients with bronchial asthma to treatment with a leukotriene receptor antagonist. Eur J Clin Invest
2002;32:949-55.
[14] Torres-Galván MJ, Ortega N, Sánchez-García F, Blanco C, Carrillo T, Quiralte J.
LTC4-synthase A-444C polymorphism: lack of association with NSAID-induced isolated periorbital angioedema in a Spanish population. Ann Allergy Asthma Immunol
2001;87:506-10.
[15] Sánchez-Borges M, Acevedo N, Vergara C, Jiménez S, Zabner-Oziel P, Monzón
A, et al. The A-444C polymorphism in the leukotriene C4 synthase gene is associated with aspirin-induced urticaria. J Investig Allergol Clin Immunol 2009;19:375-82.
[16] Acevedo N, Vergara C, Mercado D, Jiménez S, Caraballo L. The A-444C polymorphism of leukotriene C4 synthase gene is associated with IgE antibodies to
Dermatophagoides pteronyssinus in a Colombian population. J Allergy Clin Immunol
2007;119(2):505-7.
[17] Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir
Crit Care Med 2006;173(4):379-85.
[18] Eskandari HG, Unal M, Oztürk OG, Vayisoğlu Y, Muşlu N. Leukotriene C4 synthase A-444C gene polymorphism in patients with allergic rhinitis. Otolaryngol
Head Neck Surg 2006;134:997-1000.
[19] Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, Higashi N, et al.
Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirininduced asthma. J Allergy Clin Immunol 2002;109(6):936-42.
[20] Sanak M, Pierzchalska M, Bazan-Socha S, Szczeklik A. Enhanced expression of the leukotriene C(4) synthase due to overactive transcription of an allelic variant associated with aspirin-intolerant asthma. Am J Respir Cell Mol Biol 2000;23:290-6.
[21] Bizzintino JA, Khoo SK, Zhang G, Martin AC, Rueter K, Geelhoed GC, et al .
Leukotriene pathway polymorphisms are associated with altered cysteinyl leukotriene production in children with acute asthma. Prostaglandins Leukot Essent Fatty Acids
2009;81:9-15.
[22] Asano K, Shiomi T, Hasegawa N, Nakamura H, Kudo H, Matsuzaki T, et al.
Leukotriene C4 synthase gene A(-444)C polymorphism and clinical response to a CYS-
LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma.
Pharmacogenetics 2002;12(7):565-70.
[23] Isidoro-García M, Sanz C, García-Solaesa V, Pascual M, Pescador DB, Lorente F et al. PTGDR gene in asthma: a functional, genetic, and epigenetic study. Allergy
2011;66(12):1553-62.
[24] Oguma T, Palmer LJ, Birben E, Sonna LA, Asano K, Lilly CM. Role of prostanoid
DP receptor variants in susceptibility to asthma. N Engl J Med 2004;351:1752-63.
[25] Sanz C, Isidoro-García M, Dávila I, Moreno E, Laffond E, Avila C, et al. Promoter genetic variants of prostanoid DP receptor (PTGDR) gene in patients with asthma.
Allergy 2006;61:543-8.
[26] Kim SH, Kim YK, Park HW, Jee YK, Kim SH, Bahn JW, et al. Association between polymorphisms in prostanoid receptor genes and aspirin-intolerant asthma.
Pharmacogenet Genomics 2007;17:295-304.
[27] Jinnai N, Sakagami T, Sekigawa T, Kakihara M, Nakajima T, Yoshida K, et al.
Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach. Hum Mol Genet 2004;13:3203-
17.
[28] Arriba-Méndez S, Sanz C, Isidoro-García M, Pascual M, Avila C, Dávila I, et al.
Analysis of 927T > C CYSLTR1 and -444A > C LTC4S polymorphisms in children with asthma. Allergol Immunopathol (Madr) 2008;36:259-63.
[29] Arriba-Mendez S, Sanz C, Isidoro-Garcia M, Davild I, Laffond E, Horeno E, Avila
C, Lorente F. 927T>C polymorphism of the cysteinyl-leukotriene type-1 receptor
(CYSLTR1) gene in children with asthma and atopic dermatitis. Pediatr Allergy
Immunol 2006;17:323-8.
[30] Hong X, Zhou H, Tsai HJ, Wang X, Liu X, Wang B, et al. Cysteinyl leukotriene receptor 1 gene variation and risk of asthma. Eur Respir J 2009;33:42-8.
[31] Hao L, Sayers I, Cakebread JA, Barton SJ, Beghé B, Holgate ST, et al. The cysteinyl-leukotriene type 1 receptor polymorphism 927T/C is associated with atopy severity but not with asthma. Clin Exp Allergy 2006;36:735-41.