Linkage outline

BL414 Genetics Spring 2006
Linkage and Genetic Maps Outline
February 22, 2006
In the principle of independent assortment, we saw that the two parental alleles have a 50/50 chance of being transmitted to offspring. For example, the cross of
Dd x dd gives offspring with a 50/50 chance of getting the D or d allele from one parent, and a 100% of getting d from the other parent, so they have a 50% of being Dd and a 50% chance of being dd. But when genes are located together on the same chromosome, they don’t undergo independent assortment. The result is that we see them being transmitted together more often than not.
Ch. 5.1 Linkage and Recombination
Genetic linkage is the tendency of genes located on the same chromosome to be associated in inheritance more frequently than expected from their independent assortment in meiosis. If two genes are always transmitted together, they are said to exhibit complete linkage.
Some genes are linked on the same chromosome, but do not exhibit complete linkage because of the occurrence of DNA crossover events between homologous regions of homologous chromosomes. The crossover events occur during prophase I of meiosis. This crossover is called
recombination, which involves the physical breakage of DNA in a chromosome and reforming a connection to the DNA on the homologous chromosome. The result of recombination is a new combination of alleles in the gametes, different from the arrangement on the parental chromosomes. o In Drosophila, recombination only takes place in females, not in males. In humans, males 60% of the recombination of females.
Parental types are allele arrangements which are identical to the arrangements of alleles in the parents.
Recombinant types are allele arrangements different from the parental types, due to recombination or crossovers.
The frequency of recombination is calculated as the number of offspring with recombinant allele arrangements, divided by the total number. To get a percent, multiply this frequency by 100%. A recombination frequency of < 50% means that the genes are linked. A frequency of 50%
means that the genes are unlinked and undergo independent
assortment.
Each pair of linked genes has a characteristic recombination frequency.
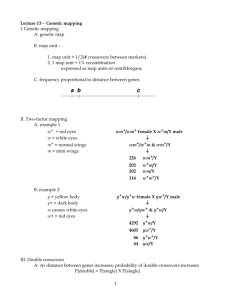
Example 1: Calculation of recombination frequencies.
P
♀w y + / w y + x
♂w + y /Y
F
1
♀w y + / w + y x ♂w y + /Y
F
2 males w y +
4484
/Y w + y /Y
4413 w + y +
76
/Y w y /Y
53 parental types:
(4484 + 4413) / 9026 x 100 = 99%
Recombinant types:
(76 + 53) / 9026 x 100 = 1%
Recombination frequency: 1%
Example 2:
Recombination frequency of w (white-eyes) and m (miniature wings) which are both X-linked.
P ♀ w+ m+ / w+ m+ x
F1 ♀ w+ m+ / w m x
♂
♂
w m / Y
w+ m+ / Y
F2 males:
412 w+ m+ / Y (parental)
389 w m / Y (parental)
206 w+ m / Y (recombinant)
185 w m+ / Y (recombinant)
Total: 1192
Recombination frequency= (206+185)/1192 x 1100% = 32.8%
Example 3:
Autosomal genes also exhibit linkage:
P ♀ b c+ / b c+ x ♂ b+ c / b+ c
F1 all b c+ / b+ c
Test cross: ♀ b c+ / b+ c x ♂ b c / b c
2934 b c+/ b c
2768 b+ c / b c
871 b c / b c
846 b+ c+ / b c
Total: 7419
Recombinant frequency= (871+846)/7419 x 100% = 23%
Terms for allele arrangement in heterozygote: BbCc o Trans, or repulsion: means that mutant genes are on opposite chromosomes: o b c+ / b+ c o Cis, or coupling: means that mutant genes are present on the same chromosome: b+ c+ / b c
Using the numbers of offspring to compare observed and expected values, the chi-square test is important for determining if recombination frequencies are significant and due to linkage or simply an effect of random sampling variation. o If the chi-square value for recombinant vs. non-recombinant offspring says the numbers are statistically significantly different, i.e. the p value < 0.05, there is linkage.
5.2 Genetic mapping
The recombination frequency is proportional to the distance between two genes on a chromosome. For short distances, 1 map unit, “m.u.” equals 1% recombination. 1 m.u. also equals 1cM, centimorgan. This is because a greater distance between two linked genes means a greater chance for a single crossover to occur between them. The frequencies can be used to build a genetic map representing the linear order of linked genes and the distances between them.
Genes located on the same chromosome are said to be syntenic, whether or not they are linked (i.e. have a recombination frequency of < 50%). A group of syntenic genes is called a linkage group. The number of linkage group for an organism is equal to the haploid number of chromosomes.
5.3 Genetic mapping in a three-point cross
Two point crosses are limited when genes are very close together or when double crossovers affect the recombination freq.
Three point crosses are more accurate in mapping genes
Example: a three point cross in corn
Looking at the linked genes:
lz: lazy or prostrate growth
gl: glossy leaf
su: sugary endosperm
A cross is done using multiply heterozygous parental genotype:
LlGgSs x LlGgSs
Progeny from a testcross of the offspring:
Phenotype of testcross progeny
Normal
Lazy
Glossy
Sugary
Lazy, glossy
Lazy, sugary
Glossy, sugary
Lazy, glossy, sugary
Genotype of gamete
Lz Gl Su lz Gl Su
Lz gl Su
Lz Gl su lz gl Su lz Gl su
Lz gl su lz gl su
Number
286
33
59
4
2
44
40
272
Total: 740
In any genetic cross involving linked genes, no matter how complex, the two most frequent types of gametes with respect to any pair of genes are nonrecombinant: these provide the linkage phase (cis vs. trans) of the alleles of the genes in the multiply heterozygous parent.
The double crossover gametes will be the least frequent types and can indicate the order of the three genes on the chromosome. A double crossover event will exchange the middle pair of alleles. This testcross suggests that the Su gene is in the middle of the other two.
Organize the data:
Parental types:
Normal Lz Su Gl
Lazy, glossy, sugary lz su gl
Single crossover between lz and su:
Glossy, sugary
Lazy, sugary
Lz su gl
Lazy lz Su Gl
Single crossover between su and gl: lz su Gl
286
272
40
33
Glossy
Double crossover types
(both lz x su and su x gl occurred)
Lz Su gl
Sugary
Lazy, glossy
Lz su Gl lz Su gl
44
2
Recombination frequency between lz and su:
(40+33+4+2)/740 = 0.107
Recombination frequency between su and gl:
(44+59+4+2)/740 = 0.147
Build the map: lz su
59
4 gl
10.7 map units
5.4 Genetic mapping in Human Pedigrees
14.7 map units
Because human pedigrees must be used to determine linkage between human genes, and the relative numbers of offspring for these will be small, statistics must be utilized to determine linkage.
The standard in human genetics is to find an lod score for a pedigree or group of pedigrees. Lod represents a “log of odds” or the logarithm of the likelihood ratio between the likelihood of linkage and the likelihood of non-linkage. A frequency of recombination, r, that maximizes the lod score is determined by iterative calculations of the lod using different r’s. The binomial coefficient is calculated for a given pedigree based on its number of outcomes (or offspring) that indicate something about recombination between the two genes of interest. An lod score
of > 3 is considered statistically significant evidence of linkage. An lod < -2 is significant evidence against linkage. A value -2 < lod < 3 is considered
uninformative and more data should be collected in order to draw a conclusion about linkage.
5.5 Tetrad Analysis
Some species of fungus create haploid spores as a product of meiosis. The genotype of these spores can be analyzed to determine linkage and gene mapping. Yeast form four spores in an unordered tetrad called an ascus.
PD: parental ditype: the alleles have the same combination found in the parents, only two different genotypes are present in the 4 spores of the tetrad.
NPD: nonparental ditype: there are two genotypes in the tetrad, and they are not in the same allele combinations as the parents
TT: tetratype tetrad: all four possible allele arrangements are present in the four spores in the tetrad