Lecture 13 - Genetic mapping I. Genetic mapping A. genetic map B
advertisement

Lecture 13 - Genetic mapping
I. Genetic mapping
A. genetic map
B. map unit –
1. map unit = 1/2(# crossovers between markers)
2. 1 map unit = 1% recombination
expressed as map units or centiMorgans
C. frequency proportional to distance between genes
II. Two-factor mapping
A. example 1
w+ = red eyes
w = white eyes
wm+/wm+ female X w+m/Y male
m+ = normal wings
m = mini wings
wm+/w+m & wm+/Y
226
wm+/Y
202
102
w+m/Y
wm/Y
114
w+m+/Y
B. example 2
y+w/y+w female X yw+/Y male
y = yellow body
y+ = dark body
y+w/yw+ & y+w/Y
w causes white eyes
w+ = red eyes
4292
y+w/Y
4605
yw+/Y
86
44
y+w+/Y
yw/Y
III. Double crossovers
A. As distance between genes increases, probability of double crossovers increases
P{double} = P{single} X P{single}
1
B. Double crossovers between markers only detected if third
gene is involved
C. as distance between markers increases, recombination
frequency provides ever increasing underestimate of genetic distance
- fairly accurate for genes relatively close together
- solution is to consider genes close together:
IV. Using recombination frequencies to order genes on chromosome
A. What is relative position of each gene?
1.
2.
3.
consider 3 X-linked genes in Drosophila
y = yellow body
rb = ruby eye color
cv = shortened wing crossvein
find recombination frequencies of:
7.5% between y & rb
6.2% between rb & cv
13.3% between y & cv
V. Three-factor mapping
A. example 1 – given gene order and genotype of parents
y = yellow body
w = white eyes
ec = echinus eye shape
y w ec/+ + + female X y w ec/Y male
4685
y w ec/y w ec & y w ec/Y
4759
+ + +/y w ec & + + +/Y
80
y + +/y w ec & y + +/Y
70
+ w ec/y w ec & y+ w ec/Y
2
(yellow, white, echinus)
(wild type)
(yellow)
(white, echinus)
193
207
3
3
10000
y w +/y w ec & y w +/Y
+ + ec/y w ec & + + ec/Y
y + ec/y w ec & y + ec/Y
+ w +/y w ec & + w +/Y
(yellow, white)
(echinus)
(yellow, echinus)
(white)
B. example 2 – don’t know whether heterozygote has all mutations in cis or trans
lz = lazy, or prostrate growth
gl = glossy leaf
su = sugary endosperm
triple heterozygote X lz gl su/lz gl su
wild type
286
lazy
33
glossy
59
sugary
4
lazy, glossy
2
lazy, sugary
44
glossy, sugary
40
lazy, glossy, sugary
272
740
C. Example 3: order and cis/trans unknown
cross triple heterozygote X homozygous recessive
wild type
black, waxy, cinnabar
waxy, cinnabar
black
cinnabar
black, waxy
waxy
black, cinnabar
5
6
69
67
382
379
48
44
1000
3
D. Points to remember about 3-factor cross
1. can be used to determine gene order and distances
2. 3-factor cross allows us to see doubles
3. double crossover switches the middle gene relative to others
4. gamete genotypes must be apparent from phenotypes of progeny
5. general strategy:
a. cross 2 true breeding parents to create triple heterozygote
b. cross triple heterozygote X homozygous recessive
c. observe F2 progeny phenotypes
d. look for parentals
e. look for products of double crossover
f. determine order from doubles
g. calculate recombination frequencies
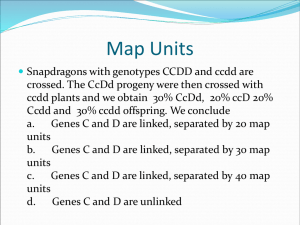
VI. Chi-square to test linkage – a statistical test for linkage
- eg. 1: Test cross het parent and find:
-
To test, we need to test the null hypothesis (that they are not linked)
2 = (observed – expected)2/expected
Need to look in table to see whether null hypothesis can be rejected
-
eg. 2: Test cross het parent and find?
- Test the null hypothesis (that they are not linked)
− χ2 = Σ(observed – expected)2/expected
4
5