File - Mrs. LeCompte
advertisement

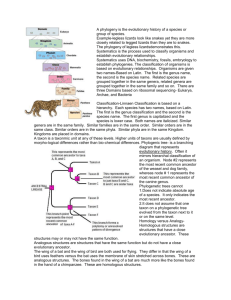
Ch. 19: Phylogeny Notes Taxonomy Developed by Linnaeus Taxon = the general name for a group containing an organism or group of organisms that exhibit a set of shared traits Phylogeny = an evolutionary family tree that represents the history of taxa Two Goals: 1) Assigning a each species with a binomial nomenclature (2-name name) called the specific epithet First name = Genus (ex. Felis) Second name = species (ex. leo) Final name: Felis leo 2) Creating a filing system to group species o Overall hierarchy Kingdom Phylum Class Order Family Genus Species The longer these match, the more closely two species are related o New evidence has led to the development of a higher taxonomic category: the Domain Due to fact that Archaea and Bacteria are less alike than previously thought, leading to the need to separate their kingdom The 3 Domain System Domain Bacteria Domain Archaea No No Membrane Lipids Phospholipids Various Branched Lipids Cell Walls with mostly… Peptidoglycan No peptidoglycan Various Extremes: Nuclear Envelopes or membrane-bound organelles? Environments HOT, SALTY or ACIDIC Domain Eukarya Includes all remaining organisms, which are eukaryotes May be multicellular or unicellular, heterotrophic or autotrophic, sexual or asexual o Multi- or uni-cellular, hetero- or autotrophic, but SIMPLE…Protists o Multicellular, AUTOtrophic…Plants o Multicellular, heterotrophic, ABSORB their food…Fungi o Multicellular, heterotrophic, INGEST their food…Animals Phylogenetic Trees In general, larger groups should break into other groups, creating finer and finer branches on the tree The Key to Classifying Organisms o Monophyletic = a single common ancestor gave rise to all species in that taxon and to no species placed in any other taxon This is the best case scenario and the goal of phylogeny o Lineage = each branch o Ancestral Traits = those found in the common ancestor and its descendants o Derived Traits = Those not found in the common ancestor (arose later) Most important for clarifying evolutionary relationships Cladistics = method of using shared, derived traits to develop a hypothesis for an evolutionary history Shown in a cladogram o Clade = a common ancestor and all it descendant lineages Applies concept of parsimony = the simplest explanation is usually correct Uses outgroup comparisons = a taxon in a lineage that does NOT have a characteristic that all the others do o Relies on information from: Fossil traits Homologous structures Must be careful of analogous traits from convergent evolution Behavioral Traits Molecular/Biochemical Evidence (DNA, RNA, or Protein comparisons) Mitochondrial DNA (mtDNA) is the best to use for closely related species, since it mutates 10x faster than nuclear DNA Can be used to create a Molecular Clock = when mutations accumulate at a fairly constant rate, they can be used to help determine a timeline of evolutionary history Ex. If two songbird subspecies have mtDNA with 5.1% differences, and we know that the average rate at which their mtDNA nucleotide changes, then we can calculate how long ago the two subspecies diverged