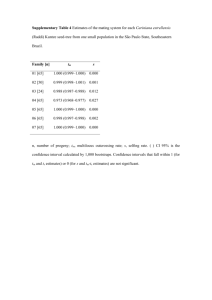
3 - Nature
advertisement

Supplementary information (Jarne & David - Quantifying inbreeding in natural populations of hermaphroditic organisms) (all in Windows Word format) Supplementary Appendix 1 (including 2 tables) – The three main molecular markers (allozymes, microsatellites and AFLPs) used for estimating the selfing rate, and associated technical problems. Supplementary Appendix 2 (including 3 figures) – The sampling properties of an estimator of the selfing rate in the single-locus case. Supplementary Appendix 3 (including 2 figures) – Joint estimation of the selfing rate and inbreeding depression. Supplementary Appendix 4 (including 1 table) – Accounting for the bias due to partial dominance when estimating the inbreeding coefficient: a general single-locus model. Supplementary Appendix 5 (including 1 figure) – Estimating the selfing rate from linkage disequilibrium data. Supplementary Appendix 6 (including 1 table and 1 figure) – The progeny-arrays approach (PAA): basic conditions and some pitfalls associated with technical problems. References (to all appendices) 2 Supplementary Appendix 1 – The three main molecular markers (allozymes, microsatellites and AFLPs) used for estimating the selfing rate, and associated technical problems. Some general characteristics of allozymes, microsatellites and AFLPs are provided in Table 1. More details are given in Avise (2000) and Lowe et al. (Lowe et al., 2004, chapter 2). These characteristics should be thoroughly considered before launching a study on selfing rates. A critical step is to produce as fair as possible data which requires using controls at several steps (Lowe et al., 2004; Hoffman and Amos, 2005; Pompanon et al., 2005). The influence of a given marker’s biological characteristics, as well as associated technical problems, on the estimation of selfing rates are also discussed in main text. The technical problems are detailed in Table 2, and their influence on the estimation of selfing rate is explained in main text, in Supplementary Appendix 4 (estimates based on the inbreeding coefficient) and in Supplementary Appendix 6 (progeny-arrays analyses). 3 Table 1. Some general characteristics of allozymes, microsatellites and AFLPs. SAD = short allele dominance; * = low; ** = intermediate; *** = high. The financial costs include development and subsequent use. The table has been built with diploid organisms in mind, and the situation is generally more complex with polyploids. a depends on the number of primer pairs used; b the Esterase family is an example; c some technical problems are presumably more acute with dinucleotide motifs than with larger motifs; d refers to erroneous reading of an allele (band); e refers to the influence of environmental (e.g., room temperature) and technical (e.g., chemicals, machines) factors on result quality; f more automatized practises lead to less direct access to primary data. 4 Marker Dominance Number of loci Number of alleles / locus Mendelian transmission Allelism (problems with) Biological material Amount State / storing Technical problems Null alleles Band stuttering Fuzzy bands SAD Misreading d Repeatability Environmental influence e Technical cost Automatization f Financial costs References Allozymes Microsatellites AFLPs codominant up to a few tens often < 5 yes with multi-genes familiesb codominant up to a few tens often < 10 yes no dominant up to a few hundredsa 2 not always when more than 2 alleles / locus g fresh / frozen ng to µg fresh / frozen / alcohol ng to µg fresh / frozen / alcohol c yes no yes irrelevant yes ** * * * * Richardson et al. (1986) Pasteur et al. (1987) yes yes yes yes yes ** ** ** ** / *** ** / *** Jarne and Lagoda (1996) Estoup and Angers (1998) Ellegren (2004) irrelevant no yes irrelevant yes * *** ** ** / *** ** Vos et al. (1995) 5 Table 2. Some technical problems encountered with the three molecular markers considered here, allozymes (Al), microsatellites (M) and AFLP. For null alleles, SAD and band stuttering, appropriate methods can be used to analyse the source and magnitude of heterozygote deficiencies (e.g. Van Oosterhout et al., 2004; David et al., 2007). Source Null alleles Definition Alleles with no electrophoretic expression / phenotype, because of failed primer amplification at M loci or no enzymatic reaction at Al loci. Homozygous individuals (say 00) do not display any patterns, and heterozygous individuals (say B0) are read as homozygous for the other allele (BB) Marker Al, M Solution Design more appropriate PCR primers (M) Short allele Preferential PCR amplification of short alleles in heterozygotes, such that heterozygotes dominance are misscored as homozygotes for the shortest allele M Manipulate PCR conditions Band stuttering Stuttered patterns at M loci results from additional PCR products which differ in size from the actual allele by even (and small) numbers of unit size (e.g., two base pairs for a dinucleotide). Heterozygotes may be misscored for homozygotes, most probably when alleles are separated by a single repeat unit. M Manipulate PCR conditions Fuzzy bands Bands (signals) of larger width than expected and blurred outlines. Might be due to too much PCR products or too active enzymatic reactions. Al, M, AFLP Manipulate PCR conditions (M, AFLP) or stop enzymatic reactions (A) Miscoring Erroneous reading of bands leading to the creation of new (imaginary) alleles or to misreading of an already-existing allele Al, M Pool alleles with similar mobilities 6 Supplementary Appendix 2 – The sampling properties of an estimator of the selfing rate in the single-locus case. Our goal is to examine the sampling properties, especially the variance, of estimators of the selfing rate derived from the inbreeding coefficient (F). Several estimators of F are available (see e.g., Curie-Cohen, 1982), but we focus on the ‘total heterozygosity’ estimator ( f in 1 Curie-Cohen, 1982) which on the whole is the least biased and exhibits the lowest variance, and is the one used in this review. Let us assume an inbred population (inbreeding coefficient F) of infinite size with no mutation, migration or selection. Assume also a locus with k codominant alleles Ai with frequency pi. n individuals are sampled. AiAi AiAj (i ≠ j) Observed number aii 2 aij Expected number ( pi2 pi (1 pi ) F )n 2 pi p j (1 F )n Genotype The ‘total heterozygosity’ estimator is defined as: Hˆ Hˆ o Fˆ e (1) Hˆ e with Hˆ o 2 aij / n is the observed frequency of heterozygotes and Hˆ e 2 pˆ i pˆ j the j i j i expected frequency under random mating. Assuming that n is large enough, it is possible to derive an approximate expression of Var ( F ) , based on the Delta method (see e.g. Appendix 1 in Lynch and Walsh, 1998) and the variances and covariance of the numerator and denominator of equation (1) (Curie-Cohen, 1982). Var ( F ) 1 F H e 1 F H e 2 1 F 2 (2 pi p j ( pi p j ) 2 ) i j nH e 2 O 1 (2). 2 n 7 When alleles are equifrequent ( pi 1 / k , i ), this simplifies to: 1 F 1 F k 1 1 F 1 H e 1 F Var ( Fˆ ) nk 1 nH e Note that Var ( Fˆ ) (3). 1 1 He when F is small. It can be shown that: n(k 1) nH e 4Var ( Fˆ ) Var ( Sˆ ) (1 Fˆ ) 4 (4) which when alleles are equifrequent gives: 1 S (2 S )2 2 S 2H e 1 S Var ( Sˆ ) 2nH e (5). The variances of the inbreeding coefficient and the selfing rate are given in Figure 1 for a three-allele locus in two contrasted situations with regard to allelic frequencies. An interesting result is that the variance of S can be quite substantial when inbreeding is limited and allelic frequencies are not balanced. The influence of gene diversity on the variance of the selfing rate can be evaluated using equations (3) and (4) (equifrequent alleles) for various values of the inbreeding coefficient (Figure 2). There is indeed a clear benefit to using polymorphic loci, especially in rather outcrossing populations. When several (L) loci are available, the inbreeding coefficient can be estimated as an average value over loci. The sampling variance of F decreases with increasing L, and so does the variance in S. However, the decrease in variance is less than linear with L when L and/or S are high. This is of limited importance for large values of S because the single-locus variance is already small (Figures 1 and 2). On the other hand, it might be asked whether the sampling variance will be more efficiently minimized by increasing either n or L when S is small. In such a situation, the population is essentially composed of two classes of individuals (selfed and outcrossed), and the frequency of selfed individuals in a sample of n individuals is a binomial variable with variance S 1 S / n . This variance does not depend on the number of 8 loci used. Another source of variance derives from determining the selfed versus outcrossed status of each individual based on their genotype. This depends on the number and genetic diversity of loci. The total variance is the sum of these two sources of variance. This is illustrated in Figure 3 in which the two sources of variance are presented as a function of S (S < 0.3) in the single-locus case. When He increases (compare the situation with He = 0.66 and 1), the part of total variance attributed to the Binomial variance increases, and it is worth increasing n. When He is low, increasing L will provide more gain than when He is high. In general it seems preferable to increase n because the total variance decreases in 1/n, while only the non-Binomial component decreases when increasing L or He. However it is sometimes less costly to score more loci than more individuals (e.g., when several loci are scored in the same electrophoresis gel or co-amplified with the same PCR mix). 9 Figure 1. Variances of the inbreeding coefficient F̂ (empty squares) and the selfing rate Ŝ (full squares) as a function of the inbreeding coefficient using equations (2) and (4) in the threeallele case. n = 100. A. p1 = 0.98, p2 = p3 = 0.01. B. p1 = p2 = 0.33, p3 = 0.34. 1A 0.16 0.12 0.08 0.04 F 0.00 0 0.2 0.4 0.6 0.8 1 1B 0.03 0.02 0.01 F 0.00 0 0.2 0.4 0.6 0.8 1 10 Figure 2. Variance of the selfing rate Ŝ (equation (4)) as a function of gene diversity (He) when the inbreeding coefficient is 0.01 (triangles), 0.2 (squares) and 0.8 (circles) – corresponding to selfing rates of 0.02, 0.33 and 0.89 respectively. n = 100. Var (S ) 2.0 1.6 1.2 0.8 0.4 He 0.0 0 0.2 0.4 0.6 0.8 1 11 Figure 3. Variance of the selfing rate Ŝ as a function of S (equations (3) and (4)) when alleles are equifrequent. The total variance is given for He = 0.66 (black triangles) and 1 (black circles), and the binomial variance (independent of He) is indicated by white squares. Sampling size is 30. Var(S) 0.08 0.06 0.04 0.02 0 S 0 0.1 0.2 0.3 12 Supplementary Appendix 3 – Joint estimation of the selfing rate and inbreeding depression. Ritland (1990) proposed to jointly estimate the selfing rate and inbreeding depression based on an experimental design in which a classical progeny-arrays (PA) analysis (Gn parents and their Gn+1 offspring) is associated with estimation of the inbreeding coefficient in Gn+1 adults. This is illustrated in Figure 1 where inbreeding is given as a function of time for adults that are partially selfing (generation n), their offspring and adults from the next generation (generation n+1). Partial selfing in Gn adults increases the inbreeding coefficient at fertilization (corresponding to the primary selfing rate). Inbreeding is then reduced by natural selection (inbreeding depression). The effect is stronger in outcrossers than in selfers (lower and upper parts of panel respectively). The adult inbreeding coefficient in successive generations might differ which constitutes a departure from inbreeding equilibrium (outcrossing situation in Figure 1). Ritland (1990) derived a simple expression relating S, F and inbreeding depression (δ) assuming mixed mating and inbreeding equilibrium: S 1 F 2 F S 1 F (1). With no depression, we have equation 1 from main text. This formula can be used to depict the relationship between the selfing rate (S) and the inbreeding coefficient (F) in adults for various values of inbreeding depression (Figure 2). It suggests that progenies should be sampled as early as possible in the life-cycle to approach to the primary selfing rate, especially when inbreeding depression is strong. 13 Figure 1. Variation of the inbreeding coefficient F as a function of time in two successive generations. Selfers and outcrossers are distinguished. Four-branch stars indicate the stage at which progenies are generally genotyped in progeny-arrays (PA) analyses. Inbreeding depression, indicated by arrows, measured through Ritland’s method (Ritland, 1990) covers the period from these stars to Gn+1 adults. For the sake of clarity, a linear decline of inbreeding depression is assumed. Fertilization Sampling stage for PA F Selfers Outcrossers Fn+1 Fn Adults Gn Juven. Gn+1 Adults Gn+1 Time 14 Figure 2. Relationship between adult inbreeding coefficient (F) and the selfing rate (S) using equation (1). The values of inbreeding depression are, from right to left, 0, 0.5, 0.75 and 0.9. S 1 0.8 0.6 0.4 0.2 F 0 0 0.2 0.4 0.6 0.8 1 15 Supplementary Appendix 4 – Accounting for the bias due to partial dominance when estimating the inbreeding coefficient: a general single-locus model. Let us assume a locus with n alleles. The allelic frequency of allele Ai is pi. The population considered is at inbreeding equilibrium with actual inbreeding coefficient F. The actual observed heterozygosity and gene diversity at this locus are Ho and He. The observed values of these three parameters are F*, Ho* and He*, and they might differ from the actual values due to various technical reasons (see main text and Supplementary Appendix 1). For parameter X, the bias ΔX is defined as X X * X . The relationship F 1 H o / H e holds for both actual and observed values, and can be used to derive the bias in F: 1 H o H o F 1 F 1 1 H e H e (1). The relative biases in observed heterozygosity H o / H o and gene diversity H e / H e depend on locus characteristics (number and frequencies of alleles) and on the kind of technical artefacts. Such artefacts can be considered as various forms of dominance. We consider a general model under which heterozygotes AiAj are read as homozygotes AiAi with probability ji and as homozygotes AjAj with probability ij (0 <ji + ij < 1). Note that this means that the observed heterozygosity will always be underestimated (negative bias). The actual and observed frequencies of genotypes AiAj are Pij and Pij* respectively. Partial dominance decreases the observed frequency of heterozygous genotypes, and Pij* Pij 1 ij ji . Summing over all heterozygotes, it can be shown that: n 1 H o Ho P n i 1 j i 1 ij Ho ij ji (2) 16 where is the average apparent loss of heterozygotes due to partial dominance. Partial dominance also modifies apparent allelic frequencies. For example, an allele i dominant over most (or all) other alleles (ji >> ij for all j) will increase in apparent frequency. The frequency variation is given by: pi 1 n Pij ij ji 2 j 1, j i (3). Equation (3) can be used to derive the bias in expected heterozygosity: n n 2 H e pi 2 pi pi i 1 i 1 (4). The first term of equation (4) represents a variance in dominance among alleles and will always be positive, while the second term represents a covariance between the average dominance level of an allele and its frequency. In most situations, this term is expected to be near zero, and the bias on gene diversity will usually be negative. Equations (2) to (4) can be used to solve equation (1), and find the deviation due to partial dominance. Because both observed and expected heterozygosities are underestimated, the two effects oppose each other when computing ΔF (equation (1)). However ΔHe < ΔHo, because (ji - ij)2 << (ji + ij), and F is therefore overestimated (ΔF > 0). This general framework allows analysing situations encountered by experimenters: - Random heterozygote loss (ji = ij = / 2 for all i,j). This will happen for example when some alleles are not amplified or loose enzymatic activity by chance. - Hierarchical dominance series (ji = if i > j, and 0 if j > i). Short-allele dominance is a slightly more general case: ij g j i , with g(x) an increasing function of x verifying 0 ≤ g(x) ≤ 1 when 0 ≤ x ≤ n-1 (i < j). This allows for various shapes of curve (e.g., linear, quadratic). Band stuttering at microsatellite loci can be modelled as: ij 0 when j = i + 1, and ij 0 otherwise. 17 - Null alleles (for all j, ij = 0 if i < n and nj = 1). By convention, all null alleles will be lumped together as allele n. Formulas for ΔHo / Ho, ΔHe / He, and ΔF are provided in Table 1 (exact formulas are given together with first-order approximations). Note that for hierarchical dominance, we consider the simple situation of equifrequent alleles. The expressions remain approximately identical when this assumption is relaxed. For null alleles, we introduced a minor correction to equations (1) to (4) to take into account the fact that null homozygotes will probably be discarded from actual datasets. Note that these formulas do not account for sampling error on allelic and genotypic frequencies. Table 1. Formulas for ΔHo / Ho (always < 0), ΔHe / He (< 0, except for random heterozygote loss), ΔF and ΔS (always > 0). pn is the frequency of null alleles. Note that the formula for kn assumes equifrequent alleles. Bias Random heterozygote loss Hierarchical dominance - - ΔHo / Ho Null alleles (1 ) ( 2 / H e F ) pn O ( pn ) 2 ΔHe / He -kn Ho (1-F) 2 0 (1 H e ) 2 p ( 2 ) n (1 ) 2 He He 2 2 (1 H e ) 2 p n O( p n ) He ΔF (1-F) (1-F) + O(2) (2 F )(1 F ) pn O( pn ) ΔS (2 S )1 S 1 (1 S ) (2 S )1 S O 2 1 (1 S ) 2 S 4 3S 1 S pn O( p 2 ) n 2 S 1 S 4 3S pn with k n (n 1 2i) i n 2 (n 1) 2 2 2 , 1 (1 F ) 2 p n (1 p n ) 1 2 p n (1 p n ) / H e and p n . (1 p n )(1 p n (1 F )) (1 p n )(1 p n (1 F )) Supplementary Appendix 5 – Estimating the selfing rate from linkage disequilibrium data. Cutter (2006) estimated the selfing rate from its long-term effect on recombination. Linkage disequilibrium can be estimated from r2, the squared correlation coefficient between pairs of nucleotidic sites. In a population at drift / recombination equilibrium: r2 1 1 4 N e ce (1) with Ne the effective population size and ce the effective recombination rate. In an inbreeding population with inbreeding coefficient F, ce c1 F . At inbreeding equilibrium, F S / 2 S . This provides an estimate of the outcrossing rate as: 1 S 1 r2 r 2 1 8 N e c 1 (2). Note that this equation does not provide meaningful estimates of S when r 2 1 1 2 N e , and negative values of 1- S can easily be obtained. 1- S is presented as a function of r2 for various values of Ne in Figure 1. It is clear that the conditions of the model will not necessarily be fulfilled. It is also likely that r2 has a large variance (Hudson, 2001). This method might though be useful in highly selfing species for which both sequence data and recombination and effective population size estimates are available. For example, Cutter (2006) used it in the nematode Caenorhabditis elegans, and returned estimates at least an order of magnitude lower than direct estimates. 20 Figure 1. The outcrossing rate (log scale) as a function of r2 using equation (2) and assuming c = 0.5. From top to bottom, Ne = 50, 1000 and 100000. 1-S 1 10-2 10-4 10-6 10-8 0 0.2 0.4 0.6 0.8 r2 1 21 Supplementary Appendix 6 – The progeny-arrays approach (PAA): basic conditions and some pitfalls associated to technical problems. 1. The basic model A detailed presentation of the basic model and some of its early extensions is given in Brown et al. (1989). We briefly review it, and present recent developments. The PAA is based on the comparison of mother and offspring genotypes. Its logical underpinnings can be exposed using the simple one-locus two-alleles case with alleles A1 and A2 in frequency p and q respectively. The expected number of offspring of each genotype is derived from the mixedmating model with selfing rate S. For example, an A1A1 mother with have progenies A1A1 with probability 1 1 S q and progenies A1A2 with probability 1 S q . The expected and observed number of offspring can then be used to build the likelihood of a given array. This might be generalized over several loci. Ritland (2002) proposed the following general formulation. Let us assume that Pklij , S is the probability of observing a progeny with genotype AkAl given parental genotype AiAj and selfing rate S. Under a mixed-mating system, the multilocus likelihood becomes: Pklij S Pklij, S 1 S Pklij,1 S loci (1). loci The likelihood of family m given parent n (genotype AiAj) is Lmn P ij N kl kl with Nkl genotypes progenies with genotype AkAl. The likelihood of the array given all parental genotypes is: Ln f n Lmn with fn the frequency of parent n in the population (which depends on allelic n frequencies). The likelihood over all arrays is given by the product over all families L Ln . Parameters (here S and allelic frequencies) are estimated by maximizing L m using classical methods (inversion of the information matrix; see e.g. Appendix 4 in Lynch 22 and Walsh (1998) for a brief introduction to ML methods). This also allows building confidence intervals and constructing tests based on likelihoods (e.g., likelihood ratio tests, Burnham and Anderson, 2002). An important point is that the expectations are derived based on several assumptions. Of importance are (see also Table 1 in main text): (i) the expected values of both S and p are uniform over mothers; (ii) segregation of alleles follows Mendelian rules, (iii) mothers and offspring genotypes are known without errors (no technical problems leading to scoring bias), and (iv) no selection occurs between fertilization and the stage at which offspring are genotyped. The latter point implies that progenies should be genotyped as early as possible in order to access to the primary selfing rate, because inbreeding depression has a strong early expression. The mother genotype should not necessarily be known, but can be inferred together with the selfing rate, provided enough offspring are screened (e.g., 15 to 20). A peculiar situation is that of gymnosperms in which mother genotypes can directly be known from the megagametophyte. Recognizing that seeds or ovules belong to a single progeny-array is straightforward in sessile, brooding organisms which include plants, fungi, and several groups of animals (e.g., cnidarians). This is not true anymore in mobile species in which newborns get away from their mother (e.g., snails). In such a situation, estimating the selfing rate in natural populations is difficult and one has to resort to more or less artificial conditions. One possibility is to collect mature individuals in natural populations, set them in controlled conditions under which offspring can be attributed to a given mother and collect their offspring (e.g., Henry et al., 2005). The inferred selfing rate is that at the stage of the lifecycle at which offspring are genotyped (see point (iv) above). If the focus is on the evolution of selfing, it might be of interest to come as close as possible to the primary selfing rate. Seeds or ovules might therefore be preferred to seedlings or juveniles. 23 The basic model has been extended in several directions which are reported in main text. The reader is referred to Ritland (2002) and Thompson and Ritland (2006), as well as to MLTR documentation (http://www.genetics.forestry.ubc.ca/ritland/). 2. Markers Individuals can be genotyped for various markers (see main text), but the most widely used have been allozymes and microsatellites (Goodwillie et al., 2005; Jarne and Auld, 2006). Dominant markers, such as AFLP, can be used, but require a much larger number of loci. The reason is that fewer situations are favourable to the detection of outcrossing events: they can be detected among the offspring of recessive homozygous mothers, while both homozygotes can be used with a two-allele codominant locus. An important question is the number of families, offspring per family and loci that should be studied. There is probably no single answer to this question, and parameters such as the actual variance in S among families or locus variability should be taken into account. The answer also depends on the model considered (e.g., effective selfing, correlated matings; see main text) and the parameters to be estimated. Ritland (1986) reports simulation results suggesting that there is little gain in using more than eight to ten offspring per family when estimating S under the mixed-mating and effective selfing models. Using highly polymorphic loci allows more precise estimates, since outcrossed events are detected with less ambiguity, and the variance of various mating system estimates decreases with the number of alleles per locus (Ritland, 1988). Such loci might though be associated with larger error rates due to technical problems (Hoffman and Amos, 2005). In the correlated-matings model, the variance of the main parameters (selfing, correlation of selfing and correlation of paternity) decreases with both the number of loci and the number of alleles per locus (Ritland, 1989, 2002). In general, K. Ritland’s simulations 24 suggest that there is little gain in using more than five to six loci when estimating mating system parameters. 3. Some pitfalls: progeny-arrays and technical artefacts The influence of partial dominance and the kind of technical artefacts mentioned in main text (e.g., null alleles) have not been worked out, although K. Ritland mentions in the most recent version of MLTR documentation (May 2004) that family estimates of selfing are sensitive to scoring errors. We do not propose a general view on this problem, but as a first approach consider a very simple situation in which S estimates might be biased by null alleles. Let us assume a progeny-array analysis in which a large number of families are assessed, as well as a large number of offspring per family (to avoid sampling variance). Individuals are genotyped at a locus with three alleles (A1, A2 and A3; A3 is a null allele). We also assume that A1 and A2 have same frequency (q / 2), and that the frequency of A3 is p. To remain close to experimental conditions, we consider that families with A3A3 (null homozygotes) mother are eliminated, as well as offspring which are either A3A3, or incompatible with their mother’s genotype. Although this might look at first glance as an unlikely situation, it should be remembered that the maternal genotype is in some studies inferred from offspring genotype (and a null allele at low frequency might well be “invisible”), or even worse corrected to be consistent with those of offspring (K. Ritland, pers. comm.). This also means that the apparent allelic frequency (estimated without taking the null allele into account) of both A1 and A2 is ½. The population has inbreeding coefficient F and selfing rate S. Let Z be the apparent selfing rate. Four situations can be distinguished with regard to the mother genotypes: null homozygotes, null heterozygotes, homozygotes for A1 or A2, A1A2 heterozygotes. This stratification can be used to derive LZ / M , O , the likelihood of Z given the mother and 25 offspring genotypes. The likelihoods are given in Table 1, together with the expected frequencies of mothers and their actual and scored genotypes. The log-likelihood of the whole sample can be derived from Table 1 (removing a constant and taking into account that p* = 0 and q* = ½ with p* and q* the frequencies of A1 and A2 estimated on data): q q LZ / M , O ln 1 Z P1 1 1 S P2 1 1 S 2 4 t 2 q q S q ln 1 Z P1 1 S P2 1 S ln 2 P3 1 S p 1 S 2 4t 2 2 2 (2). This can more simply be written as: LZ / M , O ln 1 Z Q1 ln 1 Z Q2 c , where Q1 and Q2 correspond to the first and second terms in equation (2), and c is a constant with regard to Z (third term). The maximum likelihood value of Z can be found by deriving this equation with regard to Z and equating to 0. It comes Z Q1 Q2 /Q1 Q2 , or: Z S 1 S 8 5S p 2(S 1)2 2S 5 p 2 4(S 1)3 S 2 p3 Op 4 32 S 9S 2 27S 2 (3). When S is small, Z 4 p 3 . This might be compared to the situation when the selfing rate is estimated using the inbreeding coefficient (Supplementary Appendix 4) in which the bias is of order 4p. The difference between actual and estimated selfing rates increases with the null allele frequency and decreases with the selfing rate. An illustration is provided in Figure 1. As mentioned in main text, technical problems can be detected when genotyping a large enough number of progenies. Table 1. Mother and offspring genotypes at a locus with three alleles, together with frequencies and likelihoods ( LZ / M , O ).A3 is a null allele, and the actual genotypes might differ from the scored genotypes. Null homozygous mother and offspring, as well as incompatible motheroffspring pairs, are discarded (grey overlay). Frequencies are given assuming that A3 is a regular allele, and likelihoods assuming that it is a null allele (denoted 0). The frequencies of A1, A2 and A3 are q/2, q/2 and p. S is the actual selfing rate, and Z the selfing rate to be estimated taking into account the occurrence of a null allele. p* and q* are frequency estimates from data, i.e. 0 and ½ respectively. 2 2 qF 1 F q 2 2 pq1 F q1 F , Q pF 1 F p , P2 and P3 . In the second and third rows, mother P1 t2 3S 1 S 3 p 4 , 1 Q 1 Q 2(1 Q) genotypes before (resp. after) “/” are associated to offspring genotypes before (resp. after) “/”. More details in text. 27 Actual genot. A3A3 A1A1/ A2A2 A1A3 / A2A3 A1A2 Mother Scored genot. 00 A1A1 / A2A2 A1A1 / A2A2 A1A2 Freq. Q P1 P2 P3 LZ / M , O A1A3 or A2A3 Offspring Scored genot. Frequency 00 S 1 S p A1A1 or A2A2 1 S q 2 1 Z q* A1A1 or A2A2 A1A2 A1A1 / A2A2 A2A2 / A1A1 A1A2 / A1A2 A1A1 or A2A2 A1A2 A1A1 / A2A2 A2A2 / A1A1 A1A2 / A1A2 0 0 Z 1 Z q* 1 Z 2 0 1 Z q* 1 Z 2 A1A3 / A2A3 A1A1 / A2A2 A2A3 /A1A3 A3A3 / A3A3 A3A3 / A3A3 A1A3 / A2A3 A2A3 /A1A3 A1A2 / A1A2 A1A1 / A2A2 A2A2 / A1A1 A3A3 A1A3 or A2A3 A1A1 or A2A2 A2A2 / A1A1 00 / 00 00 / 00 A1A1 / A2A2 A2A2 / A1A1 A1A2 / A1A2 A1A1/ A2A2 A2A2 / A1A1 00 A1A1 or A2A2 A1A1 or A2A2 A1A2 A1A2 Actual genot. A3A3 0 0 S 1 S q 2 0 1 S q 2 1 S p 0 0 excluded S (1 S )1 q 2 2t2 excluded 1 S q 4t2 S 1 S q 4t2 0 0 1 S p S 2 1 S q 2 S 2 1 S q 2 Z 1 Z p* 2 Z 1 Z q* 1 Z 2 0 0 0 Z 1 Z q* 1 Z 2 0 1 Z q* 1 Z 2 Z 1 Z q 1 Z 2 * 0 0 Z 4 1 Z q* 2 1 4 Z 4 1 Z q* 2 1 4 Z 2 1 Z q* 1 2 Figure 1. Difference between the estimated and actual selfing rates (ΔS) as a function of the null allele frequency (p) for various values of S (diamonds: 0; squares: 0.2; triangles: 0.5; crosses: 0.8) in the single-locus PAA. The difference is given by equation (3). ΔS 0.5 0.4 0.3 0.2 0.1 p 0 0 0.1 0.2 0.3 29 References to Supplementary information Avise JC (2000). Phylogeography. Harvard University Press: Cambridge, Massachusetts. Brown AHD, Buron JJ, Jarosz AM (1989). Isozyme analysis of plant mating systems. In Soltis D, Soltis P (eds) Isozymes in plant biology, Dioscorides Press. Pp. 73-86. Burnham KP, Anderson DR (2002). Model selection and multimodel inference: a practical information-theoretic approach. Springer-Verlag: New York. Curie-Cohen M (1982). Estimates of inbreeding in a natural population: a comparison of sampling properties. Genetics 100: 339-358. Cutter AD (2006). Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172: 171-184. David P, Pujol B, Viard F, Castella E, Goudet J (2007). Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 16: 2474-2487. Ellegren H (2004). Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 5: 435-445. Estoup A, Angers B (1998). Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In Carvalho G (eds) Advances in Molecular Ecology, NATO press: Amsterdam. Pp. 55-86. Goodwillie C, Kalisz S, Eckert CG (2005). The evolutionary enigma of mixed mating in plants: occurrence, theoretical explanations, and empirical evidence. Ann. Rev. Ecol. Evol. Syst. 36: 47–79. Henry P-Y, Bousset L, Sourrouille P, Jarne P (2005). Partial selfing, ecological disturbance and reproductive assurance in an invasive freshwater snail. Heredity 95: 428-436. Hoffman JI, Amos W (2005). Micosatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol. Ecol. 14: 599-612. 30 Hudson RR (2001). Two-locus sampling distributions and their application. Genetics 159: 1805-1817. Jarne P, Auld JR (2006). Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 60: 1816-1824. Jarne P, Lagoda PJL (1996). Microsatellites, from molecules to populations and back. Tr. Ecol. Evol. 11: 424-429. Lowe A, Harris S, Ashton P (2004). Ecological genetics - Design, analysis and application. Blackwell. Lynch M, Walsh B (1998). Genetics and analysis of quantitative traits. Sinauer: Sunderland, Massachusetts. Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J (1987). Manuel technique de génétique par électrophorèse des protéines. Lavoisier: Paris. Pompanon F, Bonin A, Bellemain E, Taberlet P (2005). Genotyping errors: causes, consequences and solutions. Nat. Rev. Genet. 6: 847-859. Richardson BJ, Baverstock PR, Adams M (1986). Allozyme electrophoresis: a handbook for animal systematics and population studies. Academic Press: Sidney. Ritland K (1986). Joint maximum-likelihood-estimation of genetic and mating structure using open-pollinated progenies. Biometrics 42: 25-43. Ritland K (1988). The genetic-mating structure of subdivided populations. 2. Correlated mating models. Theor. Pop. Biol. 34: 320-346. Ritland K (1989). Correlated matings in the partial selfer Mimulus guttatus. Evolution 43: 848-859. Ritland K (1990). Inferences about inbreeding depression based on changes of the inbreeding coefficient. Evolution 44: 1230-1241. 31 Ritland K (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity 88: 221-228. Thompson SL, Ritland K (2006). A novel mating system analysis for modes of self-oriented mating applied to diploid and polyploid arctic Easter daisies (Townsendia hookeri). Heredity 97: 119-126. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. 4: 535-538. Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M et al. (1995). AFLP- A new technique for DNA-fingerprinting. Nucl. Ac. Res. 23: 4407-4414.