Chromatin Immunoprecipitation Assay: After stimulation
advertisement

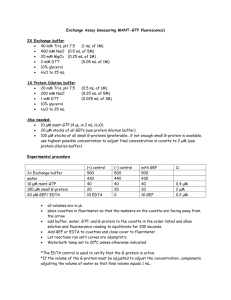
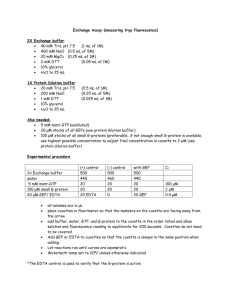
Supplementary data 6 Supplement 6: Chromatin Immunoprecipitation Assay: After stimulation, BMM were fixed in formaldehyde (HCHO; from a 37% HCHO/10% methanol stock; Fisher) at a final concentration of 1%. Crosslinking was stopped by the addition of glycine, pH 2.5, (125 mM final concentration) After 10 min, cells were washed with ice-cold PBS, pelleted by centrifugation then lysed for 15 min in L1 buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol) supplemented with proteases inhibitors. Resulting nuclei were pelleted at 3,000 rpm and resuspended in L2 buffer (50 mM Tris, pH 8.0, 1% SDS, 5 mM EDTA). Chromatin was sheared by sonication (4 x 30 s at 30% maximum potency). Lysate was centrifuged to pellet debris, and then diluted 1/10 times in RIPA buffer (50 mM Tris, pH 8.0, 0.5% NP-40, 0.2 M NaCl, 0.5 mM EDTA). Extracts were precleared for 15 min with 20 µl of a 50% suspension of salmon sperm–saturated protein A (SS protein A). Immunoprecipitations were carried out at 4°C overnight with primary antibodies (anti-ATF3, anti-Rel, antiAcetyl H3 or anti-acetyl H4) or without antibody (control). Immune complexes were collected with protein A and washed three times (5 min each) with high salt buffer (20 mM Tris, pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA, 500 mM NaCl) and three times with low salt buffer (1x Tris/EDTA [TE]). Immune complexes were extracted in 1x TE containing 1% SDS, and protein–DNA cross-links were reverted by heating at 65°C overnight. After proteinase K digestion (100 µg, 1 h at 50°C), DNA was extracted by phenol–chloroform-isoamyl alcohol then ethanol precipitated. About 1/20 of the immunoprecipitated DNA was used in each PCR. 24 PCR amplification was performed in a final volume of 20 L to which 2 L of DNA had been added. The number of cycles used for each primer set was 25-32, (depending on the linear range). The conditions for PCR amplification are as follows: denaturing at 95oC for 45 s, annealing for 45s, and extension at 72 oC for 2 min. The annealing temperature was 52C for both IL6 and IL12b primers. For semi-quantitative analysis a PCR was also run on genomic DNA obtained from each sample before immunoprecipitation, this is noted in each figure as “Input”. Products were run on a 1.2 % agarose gel and stained with ethidium bromide. Sequences of promoter-specific primers: IL6: Sense; TGC TCA AGT GCT GAG TCA CT Anti-sense; AGA CTC ATG GGA AAA TCC CA IL12b: Sense; TTC TGC AGT CTG TGG AAT CG Anti-sense; CAG AGG GAA TAT AGA CGT CG Antibody specificity: Specificity of the antibodies used for ChIP is essential for accurate results. Western blot analysis of each antibody was utilized to determine specificity and showed single bands in extracts derived from BMM. Furthermore, immunoprecipitation analysis on cells fixed using the ChIP protocol also showed single band specificity. 25