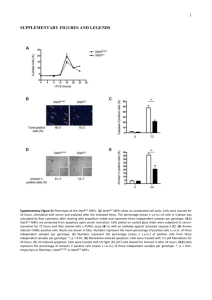
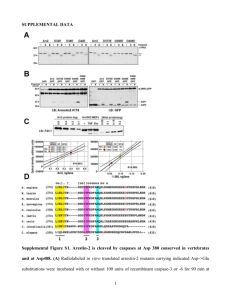
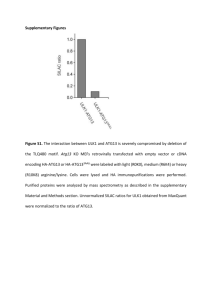
Quantification of DNA by real-time PCR:
advertisement

Supplementary Table 1 Comaparison of EGFR levels in rip1+/+/tnfr1+/+ MEFs and rip1+/+/tnfr1-/- MEFs by real-time PCR: RNA was extracted from rip1+/+/tnfr1+/+ MEFs and rip1+/+/tnfr1-/- MEFs by RNeasy Kit (Qiagen),reverse transcribed to cDNA using random primers (Invitrogen) and superscript II reverse transcriptase (Invitrogen) and quantified using real-time PCR (ABI Prism 7700, Applied Biosystems, CA). Ct values were obtained using the Sequence Detector 1.1 software. The fold difference between the two cell lines is calculated as R = 2-(delta ct cell line1-delta ct cell line2) where R is the relative expression of the gene of interest, ct is the cycle threshold, and delta ct is the difference in the ct value of the cell lines when compared with GAPDH after normalization. Each reaction was run in triplicate in the presence of SYBR-GREEN (Applied Biosystems) and repeated at least two times independently. EGFR primers used for Real Time PCR: Forward primer: AGG CAC AAG TAA CAG GCT CAC Reverse primer: AAG GTC GTA ATT CCT TTG CAC Cell line Mean SEM Normalised ct value for EGFR rip1+/+/tnfr1+/+ 22.64 3.6 MEFs rip1+/+/tnfr1-/37.30 0.17 MEFs pvalue 0.03 Results: The difference in EGFR mRNA transcript between the two cell lines was found to be 215. Thus, there is significantly more EGFR mRNA in rip1+/+/tnfr1+/+ MEFs compared to rip1+/+/tnfr1-/- MEFs.