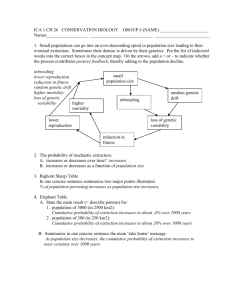
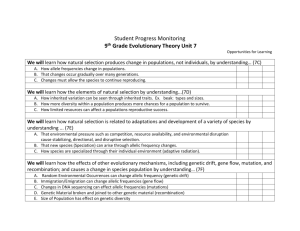
Conservation Genetics: An Historical Perspective
advertisement

Conservation Genetics: Founding Principles, Primary Concerns Ken N. Paige Foundations in Conservation Biology Jeff Brawn, Curt Meine, and Scott Robinson (Editors) University of Chicago Press List of Papers Birth of Conservation Genetics 1. Frankel, O.H. 1970. Variation, the essence of life. Sir William Macleay Memorial Lecture. Proceeding of the Linnean Society NSW 95:158-169. 2. Frankel, O.H. 1974. Genetic conservation: our evolutionary responsibility. Genetics 78:53-65. Advent of Molecular Techniques 3. Lewontin, R.C. and J.L. Hubby. 1966. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54:595609. 4. Avise, J.C., R.A. Lansman and R.O. Shade. 1979. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics 92:279-295. Loss of Genetic Diversity 5. Fisher, R.A. 1930. The Fundamental theorem of natural selection. In: The Genetical Theory of Natural Selection. Pp. 22-34. Clarendon Press, Oxford. 6. Wright, S. 1931. Evolution in mendelian populations. Genetics 16:97-159. Inbreeding and Inbreeding Depression 7. Wright, S. 1922. Coefficients of inbreeding and relationship. American Naturalist 56:330-339. 8. Davenport, C.B. 1908. Degeneration, albinism, and inbreeding. Science 28:454-455. Outbreeding Depression 9. Dobzhansky, T. 1948. Genetics of natural populations. XVIII. Experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics 33:588-602. 1 Distribution of Genetic Variation Within and Among Populations 10. Wright, S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395-420. Loss of Evolutionary Potential 11. Franklin, I.R. 1980. Evolutionary change in small populations. In Conservation Biology: an Evolutionary-Ecological Perspective (M.E. Soulé and B.A. Wilcox, editors). Pp. 135-150. Sunderland, MA, Sinauer. 12. Soulé, M.E. 1980. Thresholds for survival: maintaining fitness and evolutionary potential. In Conservation Biology: an Evolutionary-Ecological Perspective (M.E. Soulé and B.A. Wilcox, editors). Pp. 151-169. Sunderland, MA, Sinauer. 13. Lande, R. and S.J. Arnold. 1983. The measurement of selection on correlated characters. Evolution 37:1210-1226. Risk of Extinction 14. Lande, R. 1988. Genetics and demography in biological conservation. Science 241:1455-1460. 15. Gilpin, M.E. and M.E. Soulé. 1986. Minimum viable populations: processes of species extinction, In Conservation Biology: the Science of Scarcity and Diversity (M.E. Soulé, editor). Pp. 19-34. Sinauer, Sunderland, MA. Taxonomic Uncertainties and Evolutionarily Significant Units (ESUs) 16. Avise, J.C. 1989. A role for molecular genetics in the recognition and conservation of endangered species. Trends in Ecology and Evolution 4:279-281. 17. Ryder, O.A. 1986. Species conservation and systematics: the dilemma of subspecies. Trends in Ecology and Evolution 1:9-10. 2 Conservation Genetics: Founding Principles, Primary Concerns Introduction Biological diversity is rapidly being depleted due to the direct and indirect consequences of human activity. This continuing loss of populations and species is accompanied by a subtle, but no less important process--the loss of genetic diversity. When a population or species disappears all genetic information is lost as well. Furthermore, when contiguous populations are fragmented genetic diversity in the remaining fragments may diminish over time (Meffe et al. 1997). As Thomas Foose (1983) (see Temple and Fleischman section) noted, “With the accelerating destruction of the world’s wildlife and wildlands, gene pools are becoming diminished and fragmented into gene puddles”. Meffe et al. (1997) listed several biological reasons why the retention of genetic diversity is important. First, the Fundamental Theorem of Natural Selection (Fisher 1930) indicates that the rate of evolutionary change within a population is proportional to the amount of genetic variation present. In essence, the loss of genetic diversity reduces the ability of populations to evolutionarily respond to changing conditions. Second, heterozygosity is often positively correlated with fitness, given that levels of heterozygosity are directly linked to reduced population fitness through inbreeding and inbreeding depression (Reed and Frankham 2003). Third, the global pool of genetic diversity represents all information for all biological processes. Every biochemical product, every instinctive behavior, every growth pattern, every color morph is encoded in billion of bits of genetic information carried in the genome. Even epigenetic changes, heritable changes in gene activity that are not caused by changes in the DNA sequence, and consequent phenotypic plasticity may be altered by the loss of genetic diversity (Vergeer et al. 2012). The loss of such diversity will not only decrease the ability of organisms to evolutionarily respond to environmental change but will likely sacrifice useful biological information, 3 such as crop and stock genetic diversity and a suite of potentially useful, novel, and as yet untapped, products. Conservation genetics represents an integral part of the field of conservation biology, focusing on the application of genetics in conserving species as dynamic entities capable of adapting to environmental change, as well as dealing with genetic factors that affect extinction risk and genetic management regimes to minimize risk. However, as Avise (2008) pointed out, such a description may be unduly restrictive and suggests that conservation genetics be defined as the study of genetic patterns or processes in any context that informs conservation efforts. Theoretical population genetics and phylogenetics, as well as molecular genetic studies have all played key roles in the emergence of conservation genetics as a recognizable sub-discipline of conservation biology (Avise 2008). This section includes key papers and books that have been influential in the history and development of the field of conservation genetics. The foundation papers selected were published between 1908 and 1980 and cover the birth of conservation genetics, the founding principles, and some genetic aspects of the development of the field as it stands today. Why read foundational papers? For one, it is important to recognize how seminal papers have critically contributed to our understanding of processes that account for natural pattern; certainly such foundational work has irretrievably altered our perception of the ways in which the world works (Real and Brown 1991) and provided ways in which we can solve problems by applying what we have learned. Thus, to better understand where we are going it is essential to know where we have been. Furthermore, the importance of a contribution is often only apparent and appreciated in retrospect. In the case of conservation genetics, foundational research in the fields of evolutionary theory and basic population, quantitative, and molecular genetics have led to important genetic applications in mitigating 4 the loss of biodiversity and provided information important for making informed decisions in conservation biology in general. Birth of Conservation Genetics Beginning in the early 1970’s, Sir Otto Frankel was largely responsible for initial recognition of the importance of genetic factors and the role of evolution in conservation (Frankham 1995a). Until 1970, little thought was given to the genetic resources of wild biota perhaps because they were assumed to be self-renewing in natural communities (Frankel 1970, 1974). But, these communities are now disappearing at an unprecedented rate (Frankel 1974). These 1970 papers by Frankel and two additional publications in the early 1980’s, Soulé and Wilcox’s Conservation Biology: An EvolutionaryEcological Perspective (1980) and Frankel and Soulé’s Conservation and Evolution (1981), were instrumental in integrating genetics into the field of conservation biology (Schonewald-Cox 2003). With the publication of Genetics and Conservation by Schonewald-Cox, Chambers, MacBryde, and Thomas in 1983, conservation genetics was formally launched as a distinct discipline (Avise 2008). The broad topics covered in this book included the effects of isolation, extinction, bottlenecks and founder events, the natural distribution of genetic diversity, taxonomic considerations, and beginning attempts to guide management using genetic principles. Conservation genetics has rapidly matured as a science, as evidenced by the appearance of two journals over the last decade dedicated solely to conservation genetics: Conservation Genetics and Conservation Genetics Resources. Advent of Molecular Techniques The advent of molecular genetic techniques was essential in the development of the field of conservation genetics. A landmark paper by Lewontin and Hubby (1966) introduced allozyme methods to population biology, allowing for the first time, the direct measure of genetic diversity. When a mutation occurs in a gene, it can change the amino acid sequence of that gene following translation. 5 Different amino acid sequences can have different net electrical charges. Hubby and Lewontin isolated proteins from D. pseudoobscura, placed them in wells of a slab of acrylamide gel and ran a slight current across the gel. The proteins then migrated thru the gel at a speed proportional to their net electrical charge. Hubby and Lewontin observed that the same protein isolated from different members of the population frequently migrated across the gel at different speeds (represented by the "bands" in a gel), a result they correctly attributed to genetic variation. This supposition was supported by the fact that the variation segregated in a Mendelian fashion. Hubby and Lewontin's results were stunning. While some genetic variation was expected, no one was quite prepared for the enormous amounts of variation their experiments revealed (Mitton 1997, pg. 73). Hubby and Lewontin concluded that there was genetic variation at 39% of loci in the D. pseudoobscura genome. In fact, this method leads to an underestimate of the true amount of genetic variation because it does not account for mutations that do not lead to amino acid substitutions (i.e., silent mutations) or changes in the net electrical charge of the protein (Dennehy 2007). Avise, Lansman and Shade (1979), and Brown and Wright (1979) introduced the use of mitochondrial DNA methods to population biology for determining evolutionary relationships. The field has since seen an influx of new methodologies for assessing genetic diversity including restriction fragment length polymorphisms (RFLP’s), random amplified polymorphic DNA (RAPD’s), amplified fragment length polymorphisms (AFLP’s), minisatellite and microsatellite repeats, single nucleotide polymorphic markers (SNP’s), sequence data, and now expression assays (such as microarrays and RNA-seq) and proteomic and metabolomic approaches (at least for model systems) (see Avise 2004 for techniques and their applications). From a conservation genetic perspective, measures of genetic diversity for 170 threatened species examined showed that 77% had lower genetic diversity than related non-endangered species. Overall, measures of genetic diversity show that threatened species have about 6 60% of the genetic diversity of non-endangered species, primarily because threatened species have suffered reductions in population size that directly result in the loss of genetic diversity for the reasons noted below (Frankham et al. 2007). Genetic Concerns in Conservation Over the last three decades, conservation genetics has rapidly grown to encompass a suite of concerns including: 1) the loss of genetic diversity and the ability of organisms to evolutionarily respond to environmental change, including random processes such as genetic drift that override natural selection as the main evolutionary force; 2) the deleterious effects of inbreeding on reproduction and survival (i.e., inbreeding depression); 3) avoidance of deleterious effects on fitness that occur as a result of outcrossing (i.e., outbreeding depression – the breakup of co-adapted gene complexes); 4) genetic adaptation to captivity and its adverse effects on reintroduction success; 5) evolutionary responses to climate change; 6) evolutionary consequences of overexploitation and selective harvesting; and 7) resolution of taxonomic uncertainties and defining management units within species (Frankham et al. 1995a, Frankham 1999). Founding Principles of Conservation Genetics and their Application Although conservation genetics is a relatively young science, it is founded on more than a century of advances in evolutionary theory, population and quantitative genetics, and plant and animal breeding (Frankham et al. 2002). In this chapter I discuss foundational principles and how they have been applied to the issues in conservation biology listed above. Loss of Genetic Diversity in Small Populations Conservation genetics has predominantly focused on the study of small populations, in which random fluctuations in gene frequencies (random genetic drift) tend to reduce genetic diversity, leading to an increase in inbreeding and the loss of genetic diversity necessary for populations to evolutionarily 7 respond to environmental change. The mathematical treatment of genetic drift began with Fisher (1930) and Wright (1931) who considered the effects of binomial sampling in small populations. The model is often referred to as the Wright-Fisher or Fisher-Wright model (Hartl and Clark 1989). Fisher and Wright however strongly disagreed on the importance of drift in bringing about evolutionary change. Genetic drift is often called the “Sewall Wright effect” in recognition that the importance of drift in evolution was largely introduced by Wright’s arguments (Allendorf and Luikart 2007). Thus, Wright laid out the basic principles in regards to population size, drift effects and evolutionary change. In his own words Wright (1931) stated that “There remains one factor of the greatest importance in understanding the evolution of a Mendelian system. This is the size of the population. The constancy of gene frequencies in the absence of selection, mutation or migration cannot for example be expected to be absolute in populations of limited size. Merely by chance one or the other of the allelomorphs may be expected to increase its frequency in a given generation and in time the proportions may drift a long way from the original values. The decrease in heterozygosis following inbreeding is a well-known statistical consequence of such chance variation.” Wright (1931, 1969) also pointed out that the loss of genetic diversity in small populations is contingent in part upon the effective sizes (Ne) of natural populations which are usually smaller than their actual (census) sizes due to unequal numbers of males and females, fluctuations in population size, and increased variance in family size. Thus, populations will tend to lose genetic diversity much more rapidly because the “effective number” of individuals is much lower than the actual number of individuals within the population (i.e., fewer are transmitting their genetic material to the next generation). The relationship between population size, loss of genetic diversity and inbreeding in closed randomly mating populations can be described by the following equation: Ht/Ho = (1 – 1/[2Ne])t = 1 – F, where Ht is heterozygosity at generation t, Ho initial heterozygosity, Ne is the genetically effective 8 population size and F is the inbreeding coefficient, derived by Wright (1921). The equation predicts an exponential decay in genetic diversity over generations that occur at greater rates in smaller than larger populations (Frankham 2005a). Overall, only a small proportion of the genetic variation will be lost in any one generation, given that only rare alleles that contribute little to heterozygosity or heritable variation in quantitative traits are likely to be lost in a single generation of random mating (Frankel and Soulé 1981, Lande 1988). This should partially allay fears of those concerned that a single bottleneck must extract most of the genetic variation in a population (Frankel and Soulé 1981, pg. 36). The crucial issue is whether the population remains small or grows to a relatively large size; it is perennial low numbers that erode genetic variation (e.g., see Richards et al. 2003). Inbreeding and Inbreeding Depression The inbreeding coefficient equals the proportional loss in genetic diversity (or heterozygosity) (see equation above). Wright introduced the concept of the inbreeding coefficient in his 1921 paper (pg. 118) where he stated “If there is assortative mating from any cause, there will be some correlation between the gametes which unite. Represent this correlation by f.” Wright (1922) then formally defined f as the inbreeding coefficient, typically designated F. Wright (1922) also showed how to compute the inbreeding coefficient from a general pedigree by the now famous formula fo = ∑ (1/2)n + n΄ +1 (1 + fa), where summation is over all paths of length n and n΄ from the parents of O to the common ancestor A with inbreeding coefficient fa. In normally outbreeding species, inbreeding results in a decline in fitness, termed inbreeding depression. Darwin (1876) supplied the first compelling evidence for inbreeding depression from extensive plant breeding experiments, involving 57 species of plants from 52 genera and 30 families. As Darwin (1876, pg. 436) noted”…cross-fertilisation is generally beneficial, and self-fertilisation 9 injurious. This is shown by the difference in height, weight, constitutional vigour, and fertility of the offspring from crossed and self-fertilised flowers, and in the number of seeds produced by the parentplants.”; i.e., inbred plants were on average shorter, weighed less and produced fewer seeds than those that were outcrossed. The effects of inbreeding were substantial showing a 41% reduction in seed production and a 13% decline in height (Frankham et al. 2002). Since the time of Darwin, inbreeding depression has been documented in essentially all wellstudied populations of outbreeding plants and animals (Lacy 1997). Despite overwhelming evidence from laboratory (e.g., see Frankham 1995b) and domestic species (e.g., see Ralls and Ballou 1983), there was considerable skepticism regarding the occurrence of inbreeding depression in the wild (Caro and Laurenson 1994, Caughley 1994, Frankham et al. 2002). Crnokrak and Roff (1999), however, provided strong evidence that inbreeding depression commonly occurs in the wild. They reviewed 34 papers investigating inbreeding depression in the wild for 34 taxa from 157 data sets and showed that in 141 cases (90%) inbred individuals did worse than those that were outbred (Frankham et al. 2002). Two main hypotheses were formulated to account for the existence of inbreeding depression and its converse, heterosis (a gain in fitness following outbreeding) (reviewed in Wright 1977, Charlesworth and Charlesworth 1999). The overdominance hypothesis (East 1908), argues that heterozygotes are superior to each homozygote. Loss of heterozygotes through inbreeding will in turn decrease the mean value of traits associated with fitness and lead to inbreeding depression, whereas the (partial) dominance hypothesis (Davenport 1908, Jones 1917), argues that most mutations are neutral or deleterious and generally recessive (MacKay 2001). Increasing the proportion of homozygotes via inbreeding will increase the probability of unmasking these deleterious alleles (Charlesworth and Charlesworth 1999) leading to inbreeding depression (Keller & Waller 2002). Which of these two hypotheses explain most of the decline in fitness associated with inbreeding is still being debated (Karkkainen et al. 1999, Ritland 10 1996, Roff 2002). Although evidence exists to support both models (Crow 1993, Hughes 1995, Li et al. 2001, Carr and Dudash 2003, Ayroles 2009), the dominance hypothesis appears to be the favored one based on available empirical data and on theoretical grounds (Charlesworth and Charlesworth 1999). Others have suggested that inbreeding depression may also be explained, at least in part, by synergistic or epistatic interactions among genes (Templeton and Read 1994, Charlesworth 1998). Outcrossing of unrelated populations is known to reduce the detrimental effects of inbreeding, in laboratory, domestic, and wild populations of plants and animals (Frankham 2005a). Alleviation of inbreeding depression has been documented in wild populations of plants such as scarlet gilia, Ipomopsis aggregata (Heschel and Paige 1995) and white campion, Silene alba (Richards 2000), greater prairie chickens, Tympanuchus cupido (Westemeier et al. 1998), deer mice, Peromyscus maniculatus, (Schwartz and Mills 2005), gray wolves, Canis lupus (Vilà et al. 2003) and Mexican wolves, Canis lupus baileyi (Fredrickson et al. 2007), adders , Vipera berus (Madsen et al. 1999, 2004) and fish, Poeciliopsis monacha (Vrijenhoek 1994). Outbreeding Depression Outcrossing can also lead to outbreeding depression. Dobzhansky (1948) was the first to describe the phenomenon of outbreeding depression; in which sets of genes derived from different geographic populations resulted in reduced fitness when populations were brought together and bred. The underlying mechanism is either the break-up of coadapted gene complexes or genetic incompatibilities (such as chromosome number or structure causing abnormal meiosis or attendant fertility problems) (Templeton et al. 1986). Outbreeding depression is most common where there is strong genetic differentiation among populations within species or where the taxonomic status of a species is in question. For example, outbreeding has been shown to depress fitness by increasing the susceptibility of hybrid individuals and populations to infectious disease. Outbred largemouth bass 11 crossed from two geographically and genetically distinct populations suffered a 14% reduction in fitness relative to parental stocks. Furthermore, F2 generation hybrids suffered significantly higher mortality than F1 generation hybrids or wild-type parental fish following inoculation with largemouth bass virus. These results are attributed to the disruption of coadapted gene complexes in the immune system of outbred fish in the F2 generation (Goldberg et al. 2005). From a conservation perspective, outbreeding depression is particularly problematic given that wildlife managers commonly release captive-bred animals in order to maintain, rebuild or increase numbers of animals for fishing, hunting or conservation objectives (Miller et al. 2004). Nonetheless, outbreeding depression is clearly less important in animal populations than inbreeding depression given that it is often non-significant or modest in its effect (Frankham 1995a) but may be more common in plants and other organisms with low vagility (Ellstrand and Elam 1993). Even when outbreeding depression does occur it is unlikely to be a long-term problem. Hybrid populations will at worst go through a temporary decline in fitness and then increase as natural selection acts upon the extensive genetic variation in the hybrid population, leading to adaptation (Frankham et al. 2002). Distribution of Genetic Variation Within and Among Populations Wright's contributions also provided the foundation for much of the ongoing research related to population subdivision, migration and the distribution of genetic variation among population fragments (Wright 1931, 1940). Wright developed F statistics to describe the distribution of genetic diversity within and among populations (Wright 1951, 1965). In particular, the fixation index (Fst), is often used as a measure of the observed variation in allele frequencies among subpopulations. Fst is equal to 1/4Nm + 1, where N is the population size, and m the rate of migration (first described in Dobzhansky and Wright 1941). Fst is referred to as the fixation index because it increases as more subpopulations become fixed for an allele. Using F statistics Wright showed that one migrant per generation among 12 subpopulations would provide a desirable balance between genetic drift and gene flow by preventing the loss of alleles and minimizing the loss of heterozygosity while allowing genetic divergence to exist among subpopulations (Mills and Allendorf 1996). Wright, however, acknowledged that the real world complexities of social, ecological and genetic characteristics of immigrants may well require more than a single migrant per generation to prevent the random drifting apart of the genetic composition of subpopulations (Wright 1931). Nonetheless we continue to neglect these complicating factors in practice. Mills and Allendorf (1996) suggest that a minimum of 1 and a maximum of 10 migrants per generation would be an appropriate general rule of thumb, keeping in mind other factors that may influence the ideal level of connectivity among subpopulations. Vucetich and Waite (2000) concluded that more than 10 immigrants were required to compensate for increased diversification due to typical fluctuations in population size. The foundational studies of Wright (1931, 1965) also led to the underpinnings of genetic rescue in conservation (augmenting populations to alleviate the detrimental genetic effects that arise in small fragmented populations such as reduced genetic diversity and inbreeding depression). For example, Heber et al. (2013) exchanged individuals between two inbred populations of the South Island robin (Petroica australis) in New Zealand to assess whether they could restore levels of genetic diversity and alleviate inbreeding depression from these two severely inbred populations. A total of 31 individuals were exchanged (18 females from the island of Allsports to the island of Motuara and 13 females from Motuara to Allsport). Significant increases in levels of heterozygosity, allelic richness and the fixation index were observed in the following two generations, enhancing survivorship, recruitment, sperm quality, and immunocompetence. 13 Loss of Evolutionary Potential Loss of genetic diversity in small populations is also of concern due to the potential loss in the ability of populations to adapt to novel and changing environments. Wright was the first to point this out in the summary of his 1931 paper stating “In too small a population there is nearly complete fixation, little variation, little effect of selection and thus a static condition….” Monitoring genetic variation has been primarily through the use of neutral molecular markers, where heterozygosity has been the predominant and preferred measure. Neutral molecular markers are certainly useful for ascertaining pedigrees/relatedness, reconstructing phylogenies, identifying phylogeographic patterns and estimating gene flow, but molecular studies in conservation have often used them to infer adaptive features of population-genetic structure (Lynch 1996). For example, Lynch (1996) points out that a lack of molecular variation in the cheetah has been taken to infer the lack of genetic variation for adaptive evolutionary change (O’Brien et al. 1983, 1985). The rate of evolutionary change, however, is determined primarily by quantitative variation and there are several good theoretical reasons to doubt that there is a strong relationship between molecular and quantitative measures of genetic diversity. For example, the rates for recovery of quantitative variation following a population bottleneck may be higher than that for neutral molecular variation. Variation at the molecular level (heterozygosity) is introduced to a population at the per locus rate of mutation of 10-8 to 10-5 whereas quantitative variation (heritability) is introduced at the per locus rate of 10-3 to 10-2 per generation. Additional reasons why there may be no relationship between molecular and quantitative measures of diversity include nonadditive gene action, stabilizing or directional selection on quantitative traits, and sampling variance (Frankham 1999, 2005a). For example, when significant sources of variation for quantitiative traits are due to non-additive gene action (dominance and epistasis) additive genetic variance, in contrast to heterozygosity, has been observed to increase with a population bottleneck (Bryant et al. 1986, Bryant 14 and Meffert 1993, Wade et al. 1996). However, the inflation of genetic variance is typically accompanied by a reduction in fitness complicating the interpretation of its adaptive significance (Lynch 1996). The lack of a relationship between molecular and quantitative measures of genetic diversity is also borne out by a recent meta-analysis showing no statistically significant association (Reed and Frankham 2001). Fisher’s (1930) Fundamental Theorem of Natural Selection states that natural selection increases the average fitness of an organism at a rate equal to its genetic variance in fitness. That is to say that the evolutionary response depends upon the heritability which is the proportion of the variance in a trait that is due to additive genetic effects; the only component of genetic variance that can respond to selection. Thus, by extension, evolutionary potential may be reduced in threatened and endangered species given that they typically have lower levels of genetic variation than non-endangered species (Frankham 1995a, 1999, Haig and Avise 1996). Furthermore, evolutionary potential may be reduced in threatened and endangered species due to lowered reproductive success leading to lower selection differentials (approximated from the slope of the relationship between fitness and a trait), all else being equal (Frankham 1999). The response to selection can be illustrated by the breeders’ equation, R = h2S, where S is the selection differential and h2 the heritability (Falconer and Mackay 1996). The origin of the breeders’ equation is somewhat unclear, but it was clearly suggested (in multivariate form) in the early writings of Pearson and popularized by Lush (1937) (Walsh and Lynch, Evolution and Selection of Quantitative Traits, unpublished manuscript). Of course its application to evolutionary biology and by extension, conservation biology, was introduced first by Lande and Arnold in a paper published in Evolution in 1983. Heritability was first measured by Galton (1886) using parent-offspring regression (i.e., the correlation between parent and offspring). Wright (1917, 1918, 1921) followed with path analysis and Fisher (1918, 1925) developed analysis of variance techniques as ways of estimating 15 heritability. Overall, the loss of evolutionary potential will be exacerbated over time in small populations due to the continuing loss of quantitative genetic diversity and increasing homozygosity leading to further declines in reproduction, further lowering selection differentials and the ability to respond to environmental challenges. One of the more controversial areas of conservation genetics deals with how large populations should be to retain their evolutionary potential. Franklin (1980) and Soulé (1980) proposed that an effective size of 500 is sufficient for the maintenance of adequate genetic variance for adaptive evolution in quantitative traits. At equilibrium, between mutation and random genetic drift, the expected genetic variance is Vg = 2NeVm, where Vg is the additive genetic variance in a quantitative character, Ne is the effective size, and Vm is the mutational variance. Assuming a typical heritability of 0.5 where Vg = Ve, and Vm = 10-3Ve (from Lande 1975), and solving for the effective size gives an Ne of 500 (Lande 1995). More recently, Lande (1995) has argued that this number should be revised upwards, given that 90% of the mutational variance is deleterious. Thus, the Franklin-Soulé number should be increased by a factor of 10 to an effective size of 5000 (where Vm = 10-4Ve rather than Vm = 10-3Ve). Franklin and Frankham (1998), however, argued that estimates of mutational variance already include a correction for deleterious mutations, given that experiments provide an opportunity for the loss of deleterious mutations and that estimates of mutational variance in homozygous lines approximates 10-3Ve (MacKay et al. 1994). In addition, heritabilities are often less than 0.5, particularly for life history, behavioral, and physiological traits. Heritabilities most closely associated with fitness range from about 0.1 - 0.2. Thus, Ne moves back toward 500-1000 (Franklin and Frankham 1998, Frankham 1999). In response, Lynch and Lande (1998) argued that effective population sizes of 1000 or fewer could be subject to substantial random genetic drift, putting populations with low levels of genetic diversity at significant risk of extinction when adaptively challenged by changing environments. In 16 addition, the mutation rates for single-locus traits, such as disease-resistance are approximately three orders of magnitude lower than those for polygenic traits, such that much larger effective populations may be required to maintain adequate genetic diversity at such loci. Furthermore, synergistic effects between demographic, environmental, and genetic sources of stochasticity in small populations can lead to higher risks of extinction than genetic factors alone (Schultz and Lynch 1997, Lynch and Lande 1998). Lynch and Lande (1998) conclude that given the limitations in our knowledge concerning the relationships between Ne and the risk of extinction, and on deleterious versus beneficial mutations we should err on the side of caution maintaining an effective size in the range of 1000-5000. Given that the effective size of a population is often one-third to one-tenth of the actual size (Frankham 1995a, Lynch and Lande 1998), actual population size should be in excess of several thousand to maintain genetic integrity. Loss of Evolutionary Potential: Response to Climate Change One of the critical issues of 21st century conservation biology is to understand how populations will respond (and in some cases, have already responded) to climate change. The Earth’s atmosphere has warmed by approximately 0.6°C over the past 100 years and future temperature rises are likely to exceed this with a predicted rise between 0.1°C and 0.4°C per decade (e.g., see Walther et al. 2002, Jump and Penuelas 2005). Many species have already responded by extending their ranges toward the poles and some populations have been migrating, developing, or reproducing earlier in spring (Bradshaw and Holzapfel 2001). These range expansions and changes in the timing of seasonal events have generally been attributed to phenotypic plasticity (behavioral, morphological or physiological modifications in response to altered environments) (Bradshaw and Holzapfel 2006). From a conservation genetics perspective interest in rapid climate change centers on adaptive evolution, given that in the face of a long-term directional trend in the environment, evolutionary adaptation is essential 17 to population survival (i.e., if key phenotypic traits in populations are unable to adapt to rapid change the populations will likely go extinct; Burger and Lynch 1995, Conner and Hartl 2004). Recall that the rate of evolutionary change in mean phenotype (R) depends both on the strength of selection (S) and the amount of additive genetic variation (h2) (Fisher 1930). If environmental change is rapid, then this may cause rapid movement of adaptive peaks. When peaks move, populations may no longer be at an adaptive peak but rather on a slope experiencing strong directional selection. Given the likelihood of strong selection caused by environmental change, the main determinants of the ability of a population to respond to change are whether there is adequate genetic variance for rapid evolution, and how long this variance will be maintained in the face of continued strong selection. So, how can we determine which populations harbor adequate additive genetic variation to respond to environmental selection? The best approaches would be to use artificial selection (truncation selection), parent-offspring regression, or half-sibling analyses to directly measure additive genetic variation for traits needed for adapting to novel environments or common garden experiments to tease apart environmental versus genetic determinants of trait variation. However, it is often difficult to determine which traits are most important in adapting to environmental change. In addition, the large sample sizes needed for these studies and the need for controlled breeding are problematic for many rare and endangered species (Conner and Hartl 2004). Nonetheless, studies incorporating such measures have shown that over a period of decades climate change has led to heritable, genetic changes in populations of organisms as diverse as birds, mammals and insects (Bradshaw and Holzapfel 2006). For example, Canadian red squirrels faced with increasing spring temperatures and earlier spruce cone production over a ten year period were favored, via selection, to reproduce earlier. Significant changes in breeding values (representing changes in additive genetic variance and measured using restricted maximum-likelihood “animal models”; see 18 Merilä et al. 2001), in concordance with predictions from the breeder’s equation, indicated an evolutionary response to selection favoring earlier breeders (Réale et al 2003). Overall, studies providing evidence for genetic change in response to recent, rapid climate change have focused on one or a few species over several decades, involving retrospective comparisons or measures of additive genetic variation associated with responses to selection. Small organisms with short generation times and large population sizes will probably adapt to climate change and be able to maintain their populations, while many large organisms with longer generation times and small population sizes are more likely to experience declines or local extinction altogether. It is clear that unless rapid changes in climate are widely acknowledged and steps are taken to mitigate their effects, natural communities as currently recognized will cease to exist (Bradshaw and Holzapfel 2006). Risk of Extinction One of the fundamental assumptions underlying genetic concerns in conservation biology is that inbreeding and loss of genetic diversity increases the risk of extinction (Wright 1931, Frankel and Soulé 1981, Frankham 1995a, 1999) or local extirpation (assuming that there are many populations, only some of which are inbred). Wright was the first to note the risk of extinction as a result of the loss of genetic diversity in the summary of his 1931 paper where he stated “In too small a population there is nearly complete fixation, little variation ..… leading inevitably to degeneration and extinction.” While this makes intuitive sense given the relationship between the loss of genetic diversity and the ability to respond to environmental challenges, and the observed effects of inbreeding on reproductive success, evidence is limited and the issue controversial (see e.g., Caro and Laurenson 1994, Caughley 1994, Hedrick et al. 1996). In a highly influential paper, Lande (1988) ignited a debate over the importance of genetic factors in extinction, suggesting that demography and environmental stochasticity would be of more immediate importance than genetics in determining the minimum viable sizes of natural 19 populations. Furthermore, advocates of this stance (Caro and Laurenson 1994, Caughley 1994) have pointed out that the theoretical inevitability of inbreeding in small isolated populations does not necessarily translate into inbreeding depression and the likelihood of extinction (Soulé and Mills 1998). In spite of modeling results demonstrating synergistic interactions among genetic, demographic and environmental factors (Mills and Smouse 1994, Hedrick 1995), a polarization of opinion as to the relative importance of these factors persisted for more than a decade (Soulé and Mills 1998). Two studies, Saccheri et al. (1998) and Westemeier et al. (1998) brought these factors into proper perspective. Saccheri et al. (1998) provided direct evidence for the effects of inbreeding and loss of genetic diversity in extinctions of the Glanville fritillary butterfly (Melitaea cinxia) in Finland, even when demographic and environmental factors were in play. Overall, inbreeding depression explained 26% of the variation in extinction rate among butterfly populations. Similarly, a 35-year study on a remnant population of greater prairie chickens (Tympanuchus cupido) documented declines in population size and fitness as well as an overall decline in genetic diversity (Bouzat et al. 1998a,b, 2009, Westemeier et al. 1998). Translocations of individuals from large, genetically diverse populations increased fitness, due to the alleviation of inbreeding depression. The decline in demographic rates and population size occurred despite aggressive efforts in the 1960’s to the present to control predators and increase the quality and quantity of habitat (with some success). In the 1970’s however, the loss of surrounding satellite populations likely doomed the viability of the population, with nearly all breeding by greater prairie chickens in the region occurring on or within 0.8 km of managed grasslands. Once isolated, the focal population lost viability and genetic translocations became necessary (Westemeier et al. 1998). This study clearly illustrates the interactive effects of demographic and genetic events leading to the population decline, increasing the probability of extinction. Such mutually reinforcing factors are commonly known as an “extinction vortex” (Gilpin and Soule 1986). 20 Frankham and Ralls (1998) point to several lines of evidence indicating that these results likely apply to other species. Again, endangered species tend to have lower genetic diversity than nonendangered species (Frankham 1995a). This would not be expected if demographic or environmental factors drove them to extinction before genetic factors became important. Similarly, island populations often have lower genetic diversity than mainland populations with many showing inbreeding levels comparable to those of highly inbred captive populations showing fitness reductions. In addition, ratios of effective to actual population sizes appear to be much lower than previously thought, such that genetic concerns become more important in larger populations (Frankham 1995a). Furthermore, it was previously thought that purging of deleterious alleles from inbred populations would alleviate inbreeding depression. We now know from theoretical and empirical work that purging will have only modest effects in small populations, given that deleterious alleles of small effect are effectively neutral and can drift to fixation (Byers and Waller 1999, Thévenon and Couvet 2002, Reed et al. 2003). These observations and those noted above suggest that genetics can play a substantive role in the extinction process. Guidelines have recently been developed to evaluate when “genetic rescue” is a good management option (Hedrick and Fredrickson 2010). Adaptation to Captivity and Reintroductions The use of captive breeding programs is widespread and important to the conservation of many threatened and endangered species. One of the potential genetic problems associated with captive breeding is unintentional selection. Darwin (1868) was the first to point that inadvertent selection for tameness and general adaptation to captivity is inevitable in captive populations. Raising animals in benign environments may promote traits that are adaptive in captivity but maladaptive in the wild once reintroduced. A recent study by Heath et al. (2003) on chinook salmon, Oncorhynchus tshawytscha, at the Yellow Island Aquaculture hatchery in British Columbia, Canada illustrates this point. Heath et al. 21 showed that hatchery rearing relaxed natural selection that favored large eggs to the production of small eggs. The optimal egg size for hatchery females is lower than the optimal size for females in the wild resulting in lower probabilities for survival once hatchery stocks are used to supplement wild populations (egg size is strongly correlated with % egg survival). Furthermore, heavily supplemented natural populations are evolving toward smaller egg sizes, thus, gene flow is driving the evolution of suboptimal egg size in wild chinook populations. Adaptation to captivity can be minimized by equalizing family size, where differences in reproductive success can be controlled (Allendorf 1993). Equalizing family sizes will essentially act to double the effective size, diminishing the effects of selection in captivity by half (initially known as familial selection sensu Haldane 1924). The other option is to fragment populations so that genetic variation (and adaptation) within each population is reduced. Minimizing the number of generations a population is held in captivity will also reduce unintentional adaptation (Frankham 2005b, Williams and Hoffman 2009). Evolutionary Consequences of Overexploitation and Selective Harvesting Possible evolutionary consequences of harvesting have received little attention by those charged with managing wildlife populations (Rhodes and Smith 1992, Stockwell et al. 2003). The lack of attention to this issue is somewhat surprising given that harvesting can alter sex ratio, population density and age structure (e.g., Ginsberg and Milner-Gulland 1994, Solberg et al. 2000) all of which potentially influence population genetic structure (Harris et al. 2002). For example, reduced population size due to harvesting can cause a loss in genetic variation, reducing individual fitness and the ability of populations to evolve. As noted earlier, the rate of loss of heterozygosity in each generation as a result of genetic drift is measured by the effective population size (Ne). The effective population size is determined by demographic factors including the census size, generation time, sex ratio, the mean and variance in the 22 number of progeny produced by males and females, and fluctuations in population size over time (Wright 1931, 1969). Many studies report reduced genetic diversity as a result of exploitation. For example, analysis of ancient DNA shows that current populations of sea otters exhibit lower levels of heterozygosity than samples from populations predating the population bottleneck caused by fur trading in the 18th and 19th centuries (Larson et al. 2002). Similar examples (Allendorf et al. 2008; their Table 1) of reduced genetic diversity due to harvesting can be found for African elephants (Whitehouse and Harley 2001), arctic fox (Nystrom et al. 2006), New Zealand snapper (Hauser et al. 2002), sika deer (Nabata et al. 2004), tule elk (McCullough et al. 1996), red deer (Martinez et al. 2002), and white seabream (Perez-Rusafa et al. 2006). In addition, harvest often targets specific age, sex and/or size classes, thereby reducing the effective population size and increasing the rate of loss of genetic variation (Fenberg and Roy 2008). For example, using microsatellite data, Hauser et al. (2002) documented a significant decline in genetic diversity of New Zealand snapper from overharvesting larger individuals in spite of maintaining exceptionally large population sizes (in the millions), substantially reducing effective population size (Birkeland and Dayton 2005). In addition, harvesting/hunting can cause unintentional selection against the very characteristics being targeted. For example, populations of bighorn sheep are often managed to provide a source of large-horned rams for trophy hunting. In an Alberta, Canada population of bighorn sheep Coltman et al. (2003) showed an evolutionary response to sport hunting of bighorn rams in which body weight and horn size (both shown to be highly heritable traits) declined significantly over a 30 year period due to selective harvesting of the largest rams. Given that many of these harvested rams were of an average age of 6 years old and the peak age of reproduction is 8 years old, hunters imposed strong selection toward the production of smaller-horned, lighter rams and fewer trophies. Similarly, selective fishing pressure on larger/older individuals of the Northern cod caused the rapid evolution of decreased body 23 size and fecundity (Olsen et al. 2004), as life-history theory would predict (Gadgil and Bossert 1970). Thus, understanding the genetic changes and evolutionary responses of exploited populations is essential for management aimed at sustainable exploitation of wildlife species (Allendorf et al. 2008). Taxonomic Uncertainties and Evolutionarily Significant Units (ESUs) In conservation, many erroneous decisions can result if the taxonomic status of populations is not correctly assigned (Avise 1989). For example, unrecognized endangered species or subspecies may be allowed to go extinct, incorrectly classified species may be inadvertently allowed to hybridize, and populations that could be used to improve the fitness of inbred populations may be overlooked (Frankham et al. 2002). Placing several distinct species into one recognized species has resulted in the lack of protection of endangered species (Frankham et al. 2002). In the case of the tuatara, only a single species was recognized until the late 1980’s when genetic analyses (using allozymes) demonstrated that there were in fact two species, one of which was at serious risk of extinction (Daugherty et al. 1990). Hybridization between populations where taxonomic relationships were incorrect has also led to mismanagement. The last remaining dusky seaside sparrow was hybridized with the wrong seaside sparrow subspecies and became extinct (Avise and Nelson 1989). Thus, genetic data are necessary, in many (if not most) cases, along with morphological and reproductive data, to resolve taxonomic relationships. Genetic markers might include mitochondrial/chloroplast DNA (see foundational paper by Avise, Landsman and Slade 1979), microsatellites, gene sequence data, single nucleotides (SNPs) and/or allozyme data. In 1986 Ryder expressed frustration with limitations of current mammalian taxonomy in determining which named subspecies actually represent significant adaptive variation for present and future generations of the species in question and suggested that we need to identify what he termed evolutionarily significant units (ESU’s) for conservation. He recommended that concordance between 24 different data sets (e.g., range and distribution, genetics, natural history, and morphometric data) be the primary criterion for identifying ESU’s. However appealing, different data sets are often not concordant (e.g., neutral genetic markers and phenotypic traits may yield different types of evolutionary information) and would therefore create problems in making any clear management decision (Fraser and Bernatchez 2001). Waples (1991, 1995) definition of an ESU possesses the adaptive theme put forth by Ryder (1986) and is defined as a population segment or group of populations that is substantially reproductively isolated from other conspecific populations and represents an important component in the evolutionary legacy of the species. Moritz et al. (1995), among others, questioned the objectivity of Waples (1991) approach pointing out the subjectivity of the definition using words such as “substantially” and “important” making ESU designations difficult to objectively implement. Instead, Moritz (1995) favored evaluating ESU’s by emphasizing significant divergence between mitochondrial or chloroplast cytotypes, that are monophyletic within them, and nuclear loci that show significant divergence in allele frequencies (Frankham et al. 2002). The problem with this definition of ESU’s, in part, is that it ignores adaptive differentiation (Crandall et al. 2000). For example, populations with low gene flow may have differentiated by genetic drift even though they are not adaptively distinct. Crandall et al. (2000) suggest that populations be classified according to whether they show recent or historical ecological or genetic exchangeability. This would allow one to tell whether there is adaptive differentiation, gene flow, and whether gene flow is due to recent admixture or historic. Recommended management strategies are given for each of the categories of the degree of divergence designated from the data gathered (i.e., whether they are or are not ESU’s). Unfortunately, all ESU designations are flawed to some degree given that dichotomization (i.e., there is or is not an ESU) is conceptually incompatible with the continuum through which populations 25 evolve (i.e., panmixia, intermediate polyphyly, simple paraphyly, reciprocal monophyly; Omland et al. 2006, Holycross and Douglas 2007). As Holycross and Douglas (2007) point out many populations sit within that gray area in which it is equivocal whether we assign ESU status or not depending upon nuances intrinsic to the definition and criteria being used. Nonetheless, the ESU concept is currently being applied in legal and management contexts of the U.S. Endangered Species Act and the National Marine Fisheries Service (Waples 1991, 1995, Fraser and Bernatchez 2001). In practice ESU definitions appear often to be useful and include one or more of the following criteria: 1) Current geographic separation, 2) Genetic differentiation at neutral markers among related ESUs caused by past restriction of gene flow, or 3) Locally adapted phenotypic traits caused by differences in selection (Conner and Hartl 2004). For example in a study by Legge et al. (1996) Cryan’s buckmoth was evaluated for all three criteria in defining ESUs. It is found only in peatlands in the Great Lakes region of North America where they feed only on the herb buckbean, Menyanthes trifoliata. They are indistinguishable morphologically and genetically (at allozyme and mtDNA markers) from all other related buckmoths (Legge et al. 1996). However, Cryan’s buckmoth is highly adapted to its host plant with 100% survivorship, whereas two populations of close genetic relatives all died when reared on this plant. Therefore, Legge et al. (1996) concluded that Cryan’s buckmoth was an ESU based on criteria one and three, above. Thus, gene flow appears to be sufficient to reduce differentiation but not sufficient to prevent local host adaptation (Conner and Hartl 2004). Cyan’s buckmoth therefore warrants protection and management as an ESU. A New Era: Conservation Genomics Genomics will revolutionize the field of conservation genetics. The use of genome-wide markers (e.g., thousands of SNPs), for example, will substantially improve estimates of genetic and demographic parameters such as individual heterozygosity, genetic distances, measures of gene flow, 26 population structure, and kin relationships (Allendorf et al. 2010, Ouborg et al. 2010). Genomic approaches, such as transcriptomics, proteomics and metabolomics, will also undoubtedly allow the direct assessment as to how genotypic variation in populations of organisms is ultimately tied to adaptive phenotypic variation and how the loss of genetic variation affects adaptive evolutionary potential and the viability of natural populations (e.g., inbreeding depression in small populations). Developments in genomic technologies have, for example, enabled novel approaches to the study of inbreeding and inbreeding depression (Kristensen et al. 2009, Paige 2010) and selection (Kohn et al. 2006, Ouborg et al. 2010). Gene expression technology (microarray data) was used to search for transcripts directly associated with inbreeding depression for male competitive success in Drosophila melanogaster (Ayroles et al. 2009). Based on genome annotation data, it is now possible to identify genes that might be involved in a particular physiological process, such as the immune response, and as potential targets of selection. One example is the major histocompatibility complex (MHC). Studies by Hedrick et al. (2002), found several types of genomic evidence supporting the action of balancing selection in the DRB1 gene of the MHC complex in red wolves, including higher rates of nonsynonymous to synonymous substitution for the functionally important antigen-binding site positions. There is also increasing evidence that heritable variation in ecologically important phenotypic traits can be caused by epigenetic modifications of the genome, i.e., changes in gene expression caused by mechanisms other than changes in the underlying DNA sequence, such as DNA methylation and histone modifications. Such changes may provide an important and versatile mechanism for plants and animals to rapidly adapt to changing environmental conditions (Ouborg et al. 2010), perhaps in spite of reduced genetic diversity. In a recent study, Vergeer et al. (2012) compared epigenetic markers of outbred and inbred offspring of the perennial plant Scabiosa columbaria and found that inbreeding increases DNA methylation. They also experimentally demonstrated that inbreeding depression 27 disappeared when epigenetic variation was modified by treatment with a demethylation agent, linking inbreeding depression directly to epigenetic variation, suggesting an as yet unknown mechanism for inbreeding effects. Thus, the role of epigenetics will certainly be a promising future area of genomic research with relevance to the field of conservation biology. Literature Cited Allendorf, F.W. 1993. Delay of adaptation to captive breeding by equalizing family size. Conservation Biology 7:416-419. Allendorf, F.W. and G. Luikart. 2007. Conservation and the Genetics of Populations. P. 118. Blackwell Publishing, Oxford, UK. 1st edition. Allendorf, F.W., P.R. England, G. Luikart, P.A. Ritchie, and N. Ryman. 2008. Genetic effects of harvest on wild animal populations. Trends in Ecology and Evolution 23:327-337. Allendorf, F.W., P.A. Hohenlohe and G. Luikart. 2010. Genomics and the future of conservation genetics. Nature Reviews Genetics 11:697-709. Avise, J.C. 1989. A role for molecular genetics in the recognition and conservation of endangered species. Trends in Ecology and Evolution 4:279-281. Avise, J.C. 2004. Molecular Markers, Natural History, and Evolution. Sinauer Associates, Inc., Sunderland, MA, 2nd edition. Avise, J.C. 2008. The history, purview, and future of conservation genetics, In, S.P. Carroll and C.W. Fox, eds., Conservation Biology: Evolution in Action, Oxford University Press, USA. Avise, J.C., R.A. Lansman and R.O. Shade. 1979. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics 92:279-295. Avise, J.C. and W.S. Nelson. 1989. Molecular genetic relationships of the extinct dusky seaside sparrow. Science 243:646-648. Ayroles, J., K.A. Hughes, K. Rowe, M. Reedy, S. L. Rodriguez-Zas, J. Drnevich, C. Caceres, and K.N. Paige. 2009. Genome-wide assessment of inbreeding depression in Drosophila melanogaster: gene number, function and mode of action. Conservation Biology 23:920-930. Birkeland, C. and P.K. Dayton. 2005. The importance in fishery management of leaving the big ones. Trends in Ecology and Evolution 20:356-358. 28 Bouzat, J.L., H.A. Lewin, and K.N. Paige. 1998a. The ghost of genetic diversity past: ancient DNA analysis of the Greater Prairie Chicken. American Naturalist 152:1-6. Bouzat, J.L., H.H. Cheng, H.A. Lewin, R.L. Westemeier, J.D. Brawn, and K.N. Paige. 1998b. Genetic evaluation of a demographic bottleneck in the greater prairie chicken (Tympanuchus cupido). Conservation Biology 12:836-843. Bouzat, J.L., J.A. Johnson, J.E. Toepfer, S.A. Simpson, T.L. Esker and R.L. Westemeier. 2009. Beyond the beneficial effects of translocations as an effective tool for the genetic restoration of isolated populations. Conservation Genetics 10:191-201. Bradshaw, W.E. and C.M. Holzapfel. 2001. Genetic shift in photoperiodic response correlated with global warming. Proceedings of the National Academy of Sciences 98:14509-14511. Bradshaw, W.E. and C.M. Holzapfel. 2006. Evolutionary response to rapid climate change. Science 312:1477-1478. Brown, W.M. and J. Wright. 1979. Mitochondrial DNA analyses of the origin and relative age of parthenogenetic lizards (genus Cnemidophorus). Science 203:1247-1249. Bryant, E.H. and L.M. Meffert. 1993. The effect of serial founder-flush cycles on quantitative genetic variation in the housefly. Heredity 70:122-129. Bryant, E.H., L.M. Meffert and S.A. McCommas. 1986. The effect of an experimental bottleneck upon quantitative genetic variation in the housefly. Genetics 114:11911211. Burger, R. and M. Lynch. 1995. Evolution and extinction in a changing environment: A quantitative-genetic analysis. Evolution 49:151-163. Byers, D.L. and D.M. Waller. 1999. Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annual Review of Ecology and Systematics 30:479-513. Caro, T.M. and M.K. Laurenson. 1994. Ecological and genetic factors in conservation: a cautionary tale. Science 263:485-486. Carr, D.E., and M.R. Dudash. 2003. Recent approaches into the genetic basis of inbreeding depression in plants. Philosophical Transactions of the Royal Society of London Series B 358:1071-1084. Caughley, G. 1994. Directions in conservation biology. Journal of Animal Ecology 63:215-244. 29 Charlesworth, B. 1998. The effect of synergistic epistasis on the inbreeding load. Genetics Research 71:85-89. Charlesworth, B. and D. Charlesworth. 1999. The genetic basis of inbreeding depression. Genetics Research 74:329-340. Charlesworth, D., M.T. Morgan, and B. Charlesworth. 1993. Mutation accumulation in finite outbreeding and inbreeding populations. Genetical Research 61:39-56. Coltman, D.W., P. O’Donoghue, J.T. Jorgenson, J.T. Hogg, C. Strobeck, and M. Festa-Bianchet. 2003. Undesirable evolutionary consequences of trophy hunting. Nature 426:655-658. Conner, J.K. and D.L. Hartl. 2004. A Primer of Ecological Genetics. Sinauer Associates, Inc. Crandall, K.A., O.R.P. Bininda-Emonds, G.M. Mace, and R.K. Wayne. 2000. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution 15:290295. Crnokrak, P. and D.A. Roff. 1999. Inbreeding depression in the wild. Heredity 83:260-270. Crow, J.F. 1993. Mutation, mean fitness, and genetic load. Pp. 3-42, In D. Futuyma and J. Antonovics, eds. Oxford Surveys in Evolutionary Biology. Oxford Univ. Press, Oxford, U.K. Darwin, C. 1868. The variation of animals and plants under domestication, 2 volumes. London, John Murray. Darwin, C. 1876. The Effects of Cross and Self-Fertilization in the Vegetable Kingdom. John Murray, London. Davenport, C.B. 1908. Degeneration, albinism, and inbreeding. Science 28:454-455. Daugherty, C.H., A. Cree, J.M. Hay, and M.B. Thompson. 1990. Neglected taxonomy and continuing extinctions of tuatara (Sphenodon). Nature 347:177-179. Dennehy, J. 2007. The evolutionary biologist. http://evilutionarybiologist.blogspot.com/2007/08/this-weeks-citation-classic_17.html Dobzhansky, T. 1948. Genetics of natural populations. XVIII. Experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics 33:588-602. Dobzhansky, T. and S.Wright. 1941. Genetics of natural populations. V. Relations between mutation rate and accumulation of lethals in populations of Drosophila pseudoobscura. Genetics 26:23-51. 30 East, E.M. 1908. Inbreeding in corn. Report Connecticut Agricultural Experiment Station 1907: 419-428. Ellstrand, N.C. and D.R. Elam. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24:217242. Falconer, D.S. and T.F.C. MacKay. 1996. Introduction to Quantitative Genetics, 4th ed., Longman, Harlow, UK. Fenberg, P.B. and K. Roy. 2008. Ecological and evolutionary consequences of size-selective harvesting: how much do we know? Molecular Ecology 17:209-220. Fisher, R.A. 1918. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society, Edinburgh 52:399-433. Fisher, R.A. 1925. Statistical methods for research workers. Edinburgh: Oliver and Boyd. Fisher, R.A. 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. Foose, T.J. 1983. The relevance of captive populations to the conservation of biotic diversity, In Schonewald-Cox, C. et al., eds., Genetics and Conservation: A Reference for Managing Wild Animal and Plant Populations. The Benjamin-Cummings Publishing Company, Inc., Menlo Park, CA. Frankel, O.H. 1970. Variation, the essence of life. Sir William Macleay Memorial Lecture. Proceeding of the Linnean Society NSW 95:158-169 Frankel, O.H. 1974. Genetic conservation: our evolutionary responsibility. Genetics 78:53-65. Frankel, O.H. and M.E. Soulé. 1981. Conservation and Evolution. Cambridge University Press, UK. Frankham, R. 1995a. Conservation genetics. Annual Review of Genetics 29:305-327. Frankham, R. 1995b. Inbreeding and extinction: a threshold effect. Conservation Biology 9:792-799. Frankham, R. 1999. Quantitative genetics in conservation biology. Genetics Research 74:237244. Frankham, R. 2005a. Genetics and extinction. Biological Conservation 126:131-140. Frankham, R. 2005b. Stress and adaptation in conservation genetics. Journal of Evolutionary Biology 18:750-754. 31 Frankham, R., J.D. Ballou, and D.A. Briscoe. 2002. Introduction to Conservation Genetics. Cambridge University Press, UK. Frankham, R., J.D. Ballou, and D.A. Briscoe. 2007. A Primer of Conservation Genetics. Cambridge University Press, UK. Frankham, R. and K. Ralls. 1998. Conservation biology: Inbreeding leads to extinction. Nature 392:441-442. Franklin, I.R. 1980. Evolutionary change in small populations, Pp. 135-150, In, Conservation Biology: An Evolutionary-Ecological Perspective, M.E. Soulé and B.A. Wilcox, eds., Sinauer, Sunderland, MA, USA. Franklin, I.R. and R. Frankham. 1998. How large must populations be to retain evolutionary potential? Animal Conservation 1:69-70. Fraser, D.J. and L. Bernatchez. 2001. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Molecular Ecology 10:2741-2752. Fredrickson, R.J., P. Siminski, M. Woolf, and P.W. Hedrick. 2007. Genetic rescue and inbreeding depression in Mexican wolves. Proceedings of the Royal Society B. 274:2365-2371. Gadgil, M. and W. Bossert. 1970. Life history consequences of natural selection. American Naturalist 104:1-24. Gilligan, D.M., L.M. Woodworth, M.E. Montgomery, D.A. Briscoe, and R. Frankham. 1997. Is mutation accumulation a threat to the survival of endangered populations? Conservation Biology 11:1235-1241. Gilpin, M.E. and M.E. Soulé. 1986. Minimum viable populations: processes of species extinction, In Conservation Biology: the Science of Scarcity and Diversity. Pp. 19-34. Sinauer, Sunderland, MA. Ginsberg, J.R. and E.J. Milner-Gulland. 1994. Sex-biased harvesting and population dynamics in ungulates: implications for conservation and sustainable use. Conservation Biology 8:157-166. Goldberg, T.L., E.C. Grant, K.R. Inendino, T.W. Kassler, J.E. Claussen, and D.P. Philipp. 2005. Increased infectious disease susceptibility resulting from outbreeding depression. Conservation Biology 19:455-462. Haig, S.M. and J.C. Avise. 1996. Avian conservation genetics. Pp. 160-189, In, J.C. Avise and J.L. Hamrick, eds., Conservation Genetics: Case Histories from Nature. Chapman and Hall, New York, N.Y. 32 Haldane, J. B. S. 1924. A mathematical theory of natural and artificial selection. Transactions of the Cambridge Philosophical Society 23:19-41. Harris, R.B, W.A. Wal, and F.W. Allendorf. 2002. Genetic consequences of hunting: what do we know and what should we do? Wildlife Society Bulletin 30:634-643. Hartl, D.L. and A.G. Clark. 1989. Principles of population genetics. P. 66, 2nd edition, Sinauer Associates, Inc., Sunderland, Massachusetts Hauser, L., G.J. Adcock, P.J. Smith, J.H. Bernal Ramirez, and G.R. Carvalho. 2002. Loss of microsatellite diversity and low effective population size in an exploited population of New Zealand snapper (Pagrus auratus). Proceedings of the National Academy of Sciences 99:11742-11747. Heath, D.D., J.W. Heath, C.A. Bryden, R.M. Johnson, and C.W. Fox. 2003. Rapid evolution of egg size in captive salmon. Science 299:1738-1740. Heber, S., A. Varsani, S. Kuhn, A. Girg, B. Kempenaers, and J. Briskie. 2013. The genetic rescue of two bottlenecked South Island robin populations using translocations of inbred donors. Proceedings of the Royal Society B 280:1-8. Hedrick, P.W. 1995. Gene flow and genetic restoration: the Florida panther as a case study. Conservation Biology 9:996-1007. Hedrick, P.W., R.C. Lacy, F.W. Allendorf, and M.E. Soulé. 1996. Directions in conservation biology: comments on Caughley. Conservation Biology 10:13121320. Hedrick, P.W., R.N. Lee and D. Garrigan. 2002. Major histocompatibility complex variation in red wolves: evidence for common ancestry with coyotes and balancing selection. Moledcular Ecology 11:1905-1913. Hedrick, P.W. and R. Fredrickson. 2010. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conservation Genetics 11:615-626. Heschel, M.S. and K.N. Paige. 1995. Inbreeding depression, environmental stress, and population size variation in scarlet gilia (Ipomopsis aggregata). Conservation Biology 9:126-133. Holycross, A.T. and M.E. Douglas. 2007. Geographic isolation, genetic divergence, and ecological non-exchangeability define ESUs in a threatened sky-island rattlesnake. Biological Conservation 134:142-154. Hughes, K.A. 1995. The inbreeding decline and average dominance of genes acting on male lifehistory characters in Drosophila melanogaster. Genetics Research 65:41-52. 33 Jones, D.F. 1917. Dominance of linked factors as a means of accounting for heterosis. Genetics 466:465–479. Jump, A.S. and J. Penuelas. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters 8:1010-1020. Karkkainen, K., H. Kuittinen, R. van Treuren, C. Vogl, S. Oikarinen, and O. Savolainen. 1999. Genetic basis of inbreeding depression in Arabis petraea. Evolution 53:1354–1365. Keller, L. and D.M. Waller. 2002. Inbreeding effects in wild populations. Trends in Ecology and Evolution 68:252-258. Kohn, M.H., W.J. Murphy, E.A. Ostrander, and R.K. Wayne. 2006. Genomics and conservation genetics. Trends in Ecology and Evolution 21:629-637. Kristensen, T.N., K.S. Pedersen, C.J. Vermeulen and V. Loeschcke. 2009. Research in inbreeding in the ‘omic’ era. Trends in Ecology and Evolution 25:44-52. Lacy, R.C. 1997. Importance of genetic variation to the viability of mammalian populations. Journal of Mammalogy 78:320-335. Lande, R. 1975. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genetical Research 26:221-235. Lande, R. 1988. Genetics and demography in biological conservation. Science 241:1455-1460. Lande, R. 1995. Mutation and conservation. Conservation Biology 9:782-791. Lande, R. and S.J. Arnold. 1983. The measurement of selection on correlated characters. Evolution 37:1210-1226. Larson, S., R. Jameson, M. Etnier, M. Fleming, and P. Bentzen. 2002. Loss of genetic diversity in sea otters (Enhydra lutris) associated with the fur trade of the 18th and 19th centuries. Molecular Ecology 11:1899-1903. Legge, J.T., R. Roush, R. DeSalle, A.P. Vogler and B. May. 1996. Genetic criteria for establishing evolutionarily significant units in Cryan’s buckmoth. Conservation Biology 10:85-98. Lewontin, R.C. and J.L. Hubby. 1966. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54:595609. 34 Li, Z.K., L.J. Luo, H.W. Mei, D.L. Wang, Q.Y. Shu, R. Tabien, D.B. Zhong, C.S.Ying, J.W. Stansel, G.S. Khush, A.H. Paterson. 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield Genetics 158:1737-1753. Lush, J.L. 1937. Animal Breeding Plans. Iowa State College Press, Ames Iowa. Lynch, M. 1996. Quantitative genetics in conservation, In, J.C. Avise and J.L. Hamrick, eds. Conservation Genetics: Case Histories from Nature. Chapman and Hall, New York, N.Y. Lynch, M. and R. Lande. 1998. The critical effective size for a genetically secure population. Animal Conservation 1:70-72. Lynch, M. and B. Walsh. 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc., Sunderland, MA. Lynch, M., R. Bürger, D. Butcher, and W. Gabriel. 1993. The mutational meltdown in asexual populations. Journal of Heredity 84:339-344. Lynch, M., J. Conery, and R. Bürger. 1995. Mutational meltdowns in sexual populations. Evolution 49:1067-1080. MacKay, T.F.C. 2001. The genetic architecture of quantitative traits. Annual Review of Genetics 35:303–339. MacKay, T.F.C., J.D. Fry, R.F. Lyman, and S.V. Nuzhdin. 1994. Polygenic mutation in Drosophila melanogaster. Estimates from response to selection in inbred strains. Genetics 136: 937-951. Madsen, T., R. Shine, M. Olsson, and H. Wittzell. 1999. Restoration of an inbred adder population. Nature 420:34-35. Madsen, T., B. Ujvari, and M. Olsson. 2004. Novel genes continue to enhance population growth in adders (Vipera berus). Biological Conservation 120:145-147. Martinez, J.G., J. Carranza, J.L. Fernández-García, and C.B. Sánchez-Prieto. 2002. Genetic variation of red deer populations under hunting exploitation in southwestern Spain. Journal of Wildlife Management 66:1273-1282. McCullough, D.R., J.K. Fischer, and J.D. Ballou. 1996. From bottleneck to metapopulation: recovery of the tule elk in California. In: Metapopulations and Wildlife Conservation (McCullough, D.R., ed.), Pp. 375-403, Island Press. Meffe, G.K., C.R. Carroll and Contributors. 1997. Principles of Conservation Biology. Sinauer Associates, Inc., Sunderland, MA. 35 Merilä, J., B.C. Sheldon, and L.E.B. Kruuk. 2001. Explaining stasis: microevolutionary studies in natural populations. Genetica 112:199-222. Miller, L.M., T. Close, and R. Kapuscinski. 2004. Lower fitness of hatchery and hybrid rainbow trout compared to naturalized populations in Lake Superior tributaries. Molecular Ecology 13:3379-3388. Mills, L.S. and F.W. Allendorf. 1996. The one-migrant-per-generation rule in conservation and management. Conservation Biology 10:1509-1518. Mills, L.S. and P.E. Smouse. 1994. Demographic consequences of inbreeding in remnant populations. American Naturalist 144:412-431. Mitton, J.B. 1997. Selection in Natural Populations. Oxford University Press. Moritz, C. 1995. Uses of molecular phylogenies for conservation. Philosophical Transactions of the Royal Society of London Series B 349:113-118. Moritz, C., S. Lavery, and R. Slade. 1995. Using allele frequency and phylogeny to define units for conservation and management. In: Evolution and the Aquatic Ecosystem: Defining Unique Units in Population Conservation (eds Nielsen J.L., and G.A. Powers), pp.249262. Symposium 17. American Fisheries Society, Bethesda, Maryland. Nabata, D., R. Masuda, O. Takahashi, and J. Nagata. 2004. Bottleneck effects on the sika deer Cervus nippon population of Hokkaido, revealed by ancient DNA analysis. Zoological Science 21:473-481. Nyström, V., A. Angerbjörn, and L. Dalén.2006. Genetic consequences of a demographic bottleneck in the Scandinavian Arctic fox. Oikos 114:84-94. O’Brien, S.J., D.E. Wildt, D. Goldman, C.R. Merrill, and M. Bush. 1983. The cheetah is depauperate in genetic variation. Science 221:459-462. O’Brien, S.J., M.E. Roelke, L. Marker, A. Newman, C.A. Winkler, D. Meltzer, L. Colly, J.F. Evermann, M. Bush, and D.E. Wildt. 1985. Genetic basis for species vulnerability in the cheetah. Science 227:1428-1434. Olsen, E.M., M. Heino, G.R. Lilly, M.J. Morgan, J. Brattey, B. Ernande, and E. Dieckmann. 2004. Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428:932-935. Omland, K. E., J.M. Baker, and J.L. Peters. 2006. Genetic signatures of intermediate divergence: population history of Old and New World Holarctic ravens (Corvus corax). Molecular Ecology 15: 795-808. 36 Ouborg, N.J., C. Pertoldi, V. Loeschcke, R.K. Bijlsma, and P.W. Hedrick. Conservation genetics in transition to conservation genomics. Trends in Genetics 26:177-187. Paige, K.N. 2010. The functional genomics of inbreeding depression: a new approach to an old problem. 21st Century Directions in Biology. BioScience 60:267-277. Perez-Rusafa, A., M. González-Wangüemert, P. Lenfant, C. Marcos and J. A. GarciaCharton. 2006. Effects of fishing protection on the genetic structure of fish populations. Biological Conservation 129:244-255. Ralls, K., and J. Ballou. 1983. Extinction: lessons from zoos, Pp. 164-184, In Genetics and Conservation: A Reference for Managing Wild Animal and Plant Populations. Schonewald-Cox, C.M., S.M. Chambers, B. MacBryde, and W.L. Thomas, eds. The Benjamin-Cummings Publishing Company, Inc., Menlo Park, CA. Réale, D., A.G. McAdam, S. Boutin, and D. Berteaux. 2003. Genetic and plastic responses of a northern mammal to climate change. Proceedings of the Royal Society, London 270:591-596. Reed, D.H. and R. Frankham. 2001. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55:1095-1103. Reed, D.H. and R. Frankham. 2003. Population fitness is correlated with genetic diversity. Conservation Biology 17:230-237. Reed, D.H., E. Lowe, D.A. Briscoe, and R. Frankham. 2003. Inbreeding and extinction: effects of rate of inbreeding. Conservation Genetics 4:405-410. Rhodes, O.E. Jr. and M.H. Smith. 1992. Genetic perspectives in wildlife management: the case of large herbivores. In: Wildlife 2001: populations, McCullough, D.R. and R.H. Barrett, editors. Pp. 985-996. Elsevier Science, London, United Kingdom. Richards, C.M. 2000. Inbreeding depression and genetic rescue in a plant meta-population. American Naturalist 155:383-394. Richards, C.M., S.N. Emery and D.E. McCauley. 2003. Genetic and demographic dynamics of small populations of Silene latifolia. Heredity 90:181-186. Ritland, K. 1996. Inferring the genetic basis of inbreeding depression in plants. Genome 39:1-8. Roff, D.A. 2002. Inbreeding depression: tests of the overdominance and partial dominance hypotheses. Evolution 56:768-775. Ryder, O.A. 1986. Species conservation and systematics: the dilemma of subspecies. Trends in Ecology and Evolution 1:9-10. 37 Saccheri, I., M. Kuussaai, M. Kankare, P. Vikman, W. Fortelius, and I. Hanski. 1998. Inbreeding and extinction in a butterfly metapopulation. Nature 392:491- 494. Schonewald-Cox, C.M., S.M. Chambers, B. MacBryde, and W.L. Thomas. 1983. Genetics and Conservation: A Reference for Managing Wild Animal and Plant Populations. The Benjamin-Cummings Publishing Company, Inc., Menlo Park, CA. Schultz, S.T. and M. Lynch. 1997. Deleterious mutations and extinction: effects of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51:1363-1371. Schwartz, M.K. and L.S. Mills. 2005. Gene flow after inbreeding leads to higher survival in deer mice. Biological Conservation 123:413-420. Solberg, E.J., A. Loison, B.-E. Saether, and O. Strand. 2000. Age-specific harvest mortality in a Norwegian moose Alces alces population. Wildlife Biology 6:41-52. Soulé, M.E. 1980. Thresholds for survival: maintaining fitness and evolutionary potential. Pp. 151-169, In M.E. Soulé and B.A. Wilcox, Conservation Biology: An EcologicalEvolutionaryPerspective. Sinauer Associates, Inc., Sunderland, MA. Soulé, M.E. and L.S. Mills. 1998. No need to isolate genetics. Science 282:1658-1659. Soulé, M.E. and B.A. Wilcox. 1980. Conservation Biology: An Ecological-Evolutionary Perspective. Sinauer Associates, Inc., Sunderland, MA. Stockwell, C.A., A.P. Hendry, and M.T. Kinnison. 2003. Contemporary evolution meets conservation biology. Trends in Ecology and Evolution 18:94-101. Templeton, A.R. and B. Read. 1994. Inbreeding: one word, several meanings, much confusion. Pp. 91-105 in V. Loeschcke, J. Tomiuk, and S.K. Jain, eds. Conservation Genetics. Birkhäuser, Basel, Switzerland. Templeton, A.R., H. Hemmer, G. Mace, U. S. Seal, W. M. Shields, and D.S. Woodruff. 1986. Local adaptation, coadaptation, and population boundaries. Zoo Biology 5:115-125. Thévenon, S. and D. Couvet. 2002. The impact of inbreeding depression on population survival depending on demographic parameters. Animal Conservation 5:53-60. Vergeer, P., N. Wagemaker, and N. J. Ouborg. 2012. Evidence for an epigenetic role in inbreeding depression. Biology Letters 1-4. 38 Vilà, C., A-K. Sundqvist, Ö Flagstad, J. Seddon, S. Björnerfeldt, I. Kojola, A. Casulli, H. Sand, P. Wabakken and H. Ellegren. 2003. Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proceedings of the Royal Society of London Series B 270:91-97. Vrijenhoek, R.C. 1994. Genetic diversity and fitness in small populations, In, V. Loeschcke, J. Tomiuk, and S.K. Jain, eds., Conservation Genetics. Birkhäuser, Basel, Switzerland. Vucetich, J.A. and T.A. Waite. 2000. Is one migrant per generation sufficient for the genetic management of fluctuating populations? Animal Conservation 3:261-266. Wade, M.J., S.M. Schuster, and L. Stevens. 1996. Inbreeding: Its effects on response to selection for pupal weight and the heritable variance in fitness in the flour beetle, Tribolium castaneum. Evolution 50:723-733. Walther, G-R., E. Post, P. Convey, A. Menzel, C. Parmesan, T.J.C. Beebee, J-M. Fromentin, O. Hoegh-Guldberg, and F. Bairlein. 2002. Ecological responses to recent climate change. Nature 416:389-395. Waples, R.S. 1991. Pacific salmon, Oncorhynchus spp. & the definition of ‘species’ under the endangered species act. Marine Fisheries Reviews 53:11-22. Waples, R.S. 1995. Evolutionarily significant units and the conservation of biological diversity under the Endangered Species Act. In: Evolution and the Aquatic Ecosystem: Defining Unique Units in Population Conservation (J.L. Nielsen and G.A. Powers, eds.), pp.8-27. Symposium 17. American Fisheries Society, Bethesda, Maryland. Westemeier, R.L., J.D. Brawn, S.A. Simpson, T.L. Esker, R.W. Jansen, J.W. Walk, E.L. Kershner, J.L. Bouzat and K.N. Paige. 1998. Tracking the long-term decline and recovery of an isolated population. Science 282:1695-1698. Whitehouse, A.M. and E.H. Harley. 2001. Post-bottleneck genetic diversity of elephant populations in South Africa, revealed using microsatellite analysis. Molecular Ecology 10:2139-2149. Williams, S.E. and E.A. Hoffman 2009. Minimizing genetic adaptation in captive breeding programs: A review. Biological Conservation 142:2388-2400. Wright, S. 1917. The average correlation within subgroups of a population. Journal of the Washington Academy of Science 7:532-535. Wright, S. 1918. On the nature of size factors. Genetics 3:367-374. Wright, S. 1921. Systems of mating. I. The biometric relations between parents and offspring. Genetics 6:111-123. 39 Wright, S. 1922. Coefficients of inbreeding and relationship. American Naturalist 56:330-339. Wright, S. 1931. Evolution in Mendelian populations. Genetics 16:97-159. Wright, S. 1940. Breeding structure of populations in relation to speciation. American Naturalist 74:232-248. Wright, S. 1943. Isolation by distance. Genetics 28:114-138. Wright, S. 1951. The genetical structure of populations. Annals of Eugenics 15:323-354. Wright, S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395-420. Wright, S. 1969. Evolution and the Genetics of Populations. I. Genetic and biometric foundations. University of Chicago Press, Chicago. Wright, S. 1977. Evolution and the Genetics of Populations, vol. 3, Experimental results and evolutionary deductions. University of Chicago Press, Chicago, IL. Zeyl, C., M. Mizesko, J. Arjan, and G.M. Visser. 2001. Mutational meltdown in laboratory yeast populations. Evolution 55:909-917. 40