RNA Isolation – TRIzol Method
advertisement

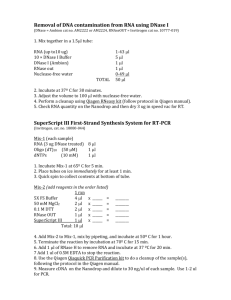
Total RNA isolation from E11.5 mouse gonads – TRIzol Method Capel Lab Protocol – Updated Jan 2009 Precautions – Work on an ice bucket unless otherwise stated in protocol. Use RNAse zap/ 70% EtOH to clean the bench before you start the protocol. Change gloves frequently to avoid contamination. Use only barrier pipette tips. Total RNA Extraction 1. Spin down gonads in RNAlater at 12000rpm for 1 minute. Remove as much RNAlater solution as you can from each tube. 2. Add 800ul TRIzol and homogenize tissue by vortexing on high for 10-20 seconds. 3. Incubate the homogenized sample for 5 minutes at 15-30C (room temp). 4. Add 160ul chlorophorm and shake tubes vigorously by hand for 15 seconds. 5. Incubate 2-3 minutes at 15-30C. 6. Spin 12,000rpm for 15 minutes at 4C. !7. CAREFULLY transfer the aqueous phase (clear phase) and put in clean tube. (Save the organic phase if isolation of DNA or protein is desired). IMPORTANT – Remove only the aqueous (clear) phase that contains the RNA. DNA is found at the interphase, and proteins are found in the organic phase. Should be able to recover about 400ul from the aqueous phase. 8. Add 5-10ug Rnase-free glycogen (~0.5-1.0 microliter, depending on concentration). 9. Add 400ul of isopropanol. 10. Incubate samples for 10 minutes at 15-30C on rocker to mix samples. 11. Spin 12,000rpm for 10 minutes at 4C. 12. Take off the supernatant carefully and discard. Should notice small RNA pellet at bottom of tube. 13. Add 800ul 75% EtOH in DEPC Water, vortex sample briefly to dislodge pellet, and spin at 10,000rpm for 5 minutes at 4C. 14. Remove EtOH and allow to dry on air for no more than 5 minutes. Pellet should stay a little damp. 15. Redissolve the RNA in Rnase-free water by passing the solution a few times through a pipette tip, and incubating for 10 minutes at 55-60C. 16. Quantify RNA concentration and quality on the Nanodrop 2000. At this point, RNA must be stored on ice or –20 to –80C at all times to minimize degradation. **If RNA sample is contaminated with phenol (260/230 < 1.7), proceed to a second extraction with chloroform. Keep in mind that some RNA will be lost during cleanup. If RNA sample looks clean, proceed to DNase digestion. RNA cleanup with additional chloroform extraction. 1. Add enough water to dissolved RNA to total 200ul. 2. Add 200ul chloroform. Shake vigorously for 15 seconds. 3. Spine 12,000rpm for 10 minutes at 4C. 4. Carefully remove top aqueous layer and put in a clean tube. Keep track of how much liquid you remove. 5. Add 3 volumes of ice-cold 100% Ethanol and mix by inverting. Add 1/10 volume of 3M NaOAc (Sodium Acetate). Add 1/100 volume of glycogen (optional). 6. Incubate at -20C for at least one hour (overnight is better but not necessary). 7. Spin at 12,000rpm for 30 minutes at 4C. 8. Discard supernatant. Add 750ul 70-75% Ethanol. Vortex briefly to dislodge pellet. 9. Spin at 12,000rpm for 15 minutes at 4C. 10. Discard supernatant. Air dry pellet. 11. Redissolve RNA in RNase-free water. Dnase Digestion For 10ul total volume: 1. Resuspend the RNA pellet in 8ul of DEPC Water (see step 15 above), then add 1ul 10x Dnase buffer and 1ul Dnase. Keep 30 minutes at 37C. 2. Add 1ul of Dnase STOP solution. Incubate at 70C for 10 minutes to stop Dnase reaction. Store RNA at -20C or -80C until needed for cDNA synthesis.