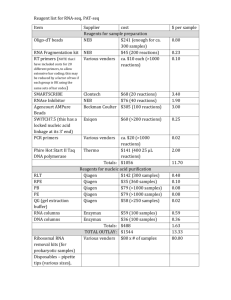
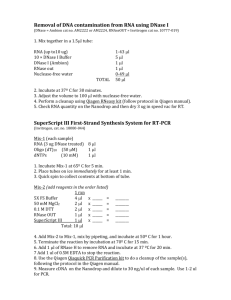
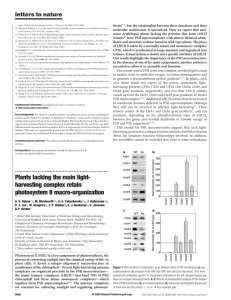
Supplementary methods Analysis of H2 and organic acid production
advertisement

Supplementary methods Analysis of H2 and organic acid production To measure H2 production, 25 L of headspace gas was withdrawn by volumetric syringe from each of six replicate vials at each time point for each control or manipulation experiment and injected directly onto a gas chromatograph with quantification of H2 by a mercuric oxide detector. To analyze organic acids, the entire liquid phase (4 mL) of each of three replicates for each time point for each control or manipulation experiment was sampled (with the associated incubation sacrificed). Liquid was filtered through 0.2 m syringe-driven filters for storage in glass vials at -20OC. Organic acids (C1-5) were quantified via high-pressure liquid chromatography (Albert and Martens, 1997). Manipulation experiments To identify the location of H2 production in microbial mats, incubations were carried out using sectioned mat cores. Mat cores were sectioned using a sterile scalpel blade yielding the following mat layers; 0-2 mm, 2-4 mm and 4-15 mm. Mat cores were sectioned and incubated under normal light conditions only and not for any of the following manipulation experiments described below. Incubations that employed addition of ammonium (NH4Cl, 8.8 mM final concentration) to field site water to suppress N2-fixation (diazotrophy) were performed to assess co-metabolic production of H2 via nitrogenases. To assess the importance of phototrophs (oxygenic and anoxygenic) in H2 production, microbial mat cores were deprived of light by wrapping aluminium foil around serum vials prior to sunrise. Assessment of the importance of oxygenic phototrophy in H2 production was carried out by inhibition of photosystem II (PSII) by addition of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU, 20 M final concentration) to field site water (Bebout et al., 1993; Oettmeier, 1992). Mat cores were incubated in DCMU amended field site water for 1 hour prior to the beginning of the incubation. To confirm that PSII was effectively inhibited using DCMU, a Walz diving-pulse amplitude modulation fluorometer (Diving PAM; Heinz Walz, Germany) was used to measure changes in PSII chlorophyll fluorescence by determination of the quantum PSII yield (PSII). Changes in PSII were analyzed using the saturation pulse method (Fleming et al., 2007; Genty et al., 1989). PSII values are relative and range from 0 or no electron transfer (inferring 0% PSII activity) to 1 or 100% electron transfer (inferring 100% PSII activity). Campbell et al. (1998) provide further information and critical reviews of this technique’s application in PSII chlorophyll fluorescence analysis of cyanobacteria. Nucleic acid isolation from microbial mats All standard precautions to mitigate against RNase contamination were employed. These precautions included: working on ice, use of and frequent changing of gloves, decontamination of surfaces and non-disposable equipment using RNase™ Zap (Ambion, TX, USA), use of disposable RNase/DNase free- plasticware and use of prepackaged RNase/DNase free filter tips. Frozen biomass cores were sectioned using a sterile scalpel blade to obtain the upper 2 mm phototrophic layer. This biomass was then transferred into a 2 ml tube containing 0.5 ml of RLT™ buffer (Qiagen, Netherlands) with 5 μl Nucleoguard™ (AmpTec, Germany). Cells within the biomass were disrupted using a rotor-stator homogenizer (Omni, GA, USA). The suspension was transferred into a new sterile 2 mL screw cap tube containing zirconium beads, and agitated for 40 sec in a FastPrep™ instrument (MP Biomedicals, OH, USA) to further disrupt cells. This mixture was centrifuged at 8,000g for 1 min to remove debris. Supernatant from at least three cores was mixed and split into separate aliquots for RNA and DNA isolation. Aliquots of supernatant for RNA isolation were processed first by adding 1 ml of acidphenol mixture. This was vortexed for 10 sec, incubated at room temperature for 5 min followed by centrifugation at 8,000g for 5 min. The aqueous phase was transferred to a new tube and passed through a 20-gauge needle 5x to shear high MW DNA. Sheared supernatant was then transferred to gDNA eliminator columns (Qiagen) and centrifuged at 8,000g for 30 sec. One volume of 80% ethanol was added to the eluate and vortexed for 10 sec. The Qiagen RNeasy Mini Kit protocol was then carried out to isolate RNA (Qiagen). Aliquots of supernatant for DNA isolation were processed by adding 1 ml of an acidphenol mixture. This was vortexed for 10 sec, incubated at room temperature for 5 min followed by centrifugation at 8,000g for 5 min. The aqueous phase was transferred to a new tube, 1 volume of 100% ethanol was added and the solution was vortexed for 10 sec. DNA was then isolated using the QIAamp DNA Mini Kit as per manufacturers protocol (Qiagen). Nucleic acids were quantified by fluorometric quantitation using the Qubit instrument (Invitrogen, CA, USA). RNA purification and cDNA synthesis Prior to cDNA synthesis, traces of contaminating DNA were removed from isolated RNA using the Turbo DNA-free kit (Ambion) to as per manufacturers protocol using two rounds of enzymatic digestion. DNA-free RNA was then further purified using the RNeasy MinElute protocol (Qiagen). RNA (1 ug) was reverse-transcribed to cDNA using random hexamers (100 ng) and the superscript III reverse transcriptase (RT) as per manufacturers protocol (Invitrogen). DNA contamination in RNA extracts was determined by performing cDNA synthesis reactions without RT. Aliquots of (-)RT reactions were then subjected to PCR in the same way as aliquots of (+)RT reactions. References Albert DB, Martens CS. (1997). Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Mar Chem 56: 2737. Bebout BM, Fitzpatrick MW, Paerl HW. (1993). Identification of the sources of energy for nitrogen fixation and physiological characterization of nitrogen-fixing members of a marine microbial mat community. Appl Environ Microbiol 59: 1495-1503. Campbell D, Hurry V, Clarke AK, Gustafsson P, Oquist G. (1998). Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol Mol Biol R 62: 667. Fleming ED, Bebout BM, Castenholz RW. (2007). Effects of salinity and light intensity on the resumption of photosynthesis in rehydrated cyanobacterial mats from Baja California Sur, Mexico. J Phycol 43: 15-24. Genty B, Briantais JM, Baker NR. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. BBAGen Subjects 990: 87-92. Oettmeier W. (1992). Herbicides of Photosystem II. In: Barber J (ed) Topics in Photosynthesis. Elsevier Science Publishers: Amsterdam, pp. 295-348.