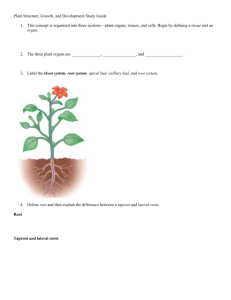
Title: Genetic approach for stem secondary growth in Arabidopsis
advertisement

Title: Genetic approach of stem secondary growth in Arabidopsis: understanding the development of wood formation. ABSTRACT Wood is an important raw material and environmentally cost-effective renewable source of energy. However, the molecular biology of wood formation (i.e. secondary growth) is surprisingly understudied. Understanding the interactions among mechanisms that regulate secondary growth will require extensive application of genomics technologies that are capable of comprehensively assaying gene expression, protein function, and metabolic pathways. Secondary growth occurs late in development and thus complicates mutant screens in more tractable species because defects in early developmental events are expected to affect secondary growth indirectly. As a result, wood formation is poorly defined at the molecular genetic level and the cambium remains the least understood plant meristem. Two developments are facilitating a paradigm shift in the study of cambium and secondary growth. First, the realization by molecular geneticists that cambium and secondary growth are not unique to threes and can be studied in the model plant Arabidopsis. Information about mechanisms that are evolutionarily conserved from model annuals, such as Arabidopsis, should be fully exploited and incorporated into studies of secondary growth in trees. The cambial activity is reduced by flowering. When it is kept from flowering by repeated removal of inflorescences, Arabidopsis produces significant quantity of secondary xylem. The later studies involving the establishment of abaxial-adaxial polarity in lateral organs, especially in leaves, have shown that three classes of genes are involved: KANADI and Class III HD-Zip. The best characterized over lap between shoot meristems and cambium regulation is seen in the class III homeodomain-leucine Zipper (HD-ZIP) and KANADI transcription factors. In Arabidopsis, KANADI and Class-III HD-ZIP genes establish the adaxial-abaxial polarity of lateral organs emerging from the shoot apical meristem. In wild-type xylem and phloem cells form on different sides of the vascular bundle. The loss-of-function mutants of adaxial-identity genes and the gain-of-function mutants of abaxial-identity genes induce amphicribal vascular bundles in which the phloem encircles the xylem. The loss-of-function mutants of abaxial-identity genes and the gain-of-function mutants of adaxial-identity genes induce amphivasal vascular bundles, whereby the xylem surrounds the phloem. In Arabidopsis, class III HD-Zip comprises five genes: PHABULOSA (PHB), PHAVOLUTA (PHAV), REVOLUTA (REV/IFL1), ATHB-8 e ATHB-15. The triple mutant rev phb phv develops radially symmetrical cotyledons with abaxialized vasculature, in which phloem develops on the adaxial and xylem on the adaxial side. In contrast to the normal collateral pattern in which phloem is on the abaxial side and xylem is on the adaxial side. Gain-of-function–PHB/AtHB14 or REV cause a dramatic transformation of abaxial-leaf fates into adaxial fates and result in the formation of an amphivasal vascular bundle in which xylem surrounds phloem. PHB, PHV and REV genes seem to regulate the cellular differentiation of the vascular system. Loss of function of rev alleles give rise to mutants with reduced number of interfasciolar xylem fibers. Double loss-of-function of rev, phb/+ and rev, phv, enhances further the mutation in the vascular system promoted by the rev mutation. This project is focused in the action of several genes belonging to these gene families during the development of secondary growth. The role that several genes belong to the CLE-family might have in the cambium, will also be studied. Genes belonging to the CLE-family are characterized for codifying for small peptides. An example of such, is the CLAVATA3 (CLV3) gene. This gene has an important role in the meristem and recent studies have shown that other genes of the CLE-family also play an important role in the proliferation of the procambium, the meristem of the vascular system. OBJECTIVES The main goal in proceeding to genetic studies of secondary growth responsible for wood formation in trees resides mainly on the fact that in Portugal wood production occupies a great portion of the territory. This economically important wood production is threatened every summer by fires. Data from 2000, show that wild fires burn each year 700 000–1 000 000 ha of wildlands in the Mediterranean basin. Reforestation with new trees that could produce more wood in a shorter period of time and requiring reduced land area, brings advantages for the ecosystems and economy. It is imperative to increase the knowledge of the integrated development of vascular plants and their vascular systems in detail. To date, the molecular studies that have been published in the last few years using whole-transcriptome profiles during wood formation in species such as Arabidopsis, aspen and pinus, has been an important contribution to elucidate the genetic mechanism of the secondary growth in trees. STATE OF THE ART Wood is the largest fraction the biomass on this planet1. In addition to the primary meristems for development of primary tissues, tree species also produce secondary tissue (wood) from the vascular cambium in a process called secondary growth, consisting of successive addition of secondary xylem2-3 which is responsible for radial expansion of woody stems. Several zones can be observed according to function in a stem cross section4. The pith is central with adjacent xylem (wood), composed mainly of dead cells involved in the transit of water and minerals5. Bark makes up the outer zone, with adjacent phloem, where sap is transported. Between the xylem and the phloem is the vascular cambium6. Thus from the center to the periphery is the pith, wood (xylem), cambium, phloem and bark. The cambial meristem produces both new xylem and phloem in a bi-directional manner, which in time form the wood and bark, respectively. Differentiation of xylem cells from procambial cells is coupled with expression of Class III HD-ZIP genes. In Arabidopsis, this gene family contains AtHB8 and AtHB15 that are expressed in vascular bundles, in particular, in the procambial or xylem precursor region10-11. Vascular bundles present in many plant leaves show a distinct dorsoventral organization - xylem is located on the dorsal (adaxial) side, phloem is on the ventral (abaxial) side with procambium positioned between xylem and phloem. Antirrhinum majus and A. thaliana mutants that show defects in the identity or maintenance of the dorsoventral axis are indicative of the involvement of the dorsoventral axis in the radialpattern formation of leaf vascular tissues7-9. There is a relationship between vascular and overall body patterning as mutations disrupting directional growth such as establishment of new growth axes, usually, also interfere with vascular strand formation. For example, the Arabidopsis triple mutant of the Class III HD-ZIP genes; rev phb phv, develops radially symmetrical cotyledons with abaxialized vasculature, in which phloem develops on the adaxial and xylem on the adaxial side12. In contrast to the normal collateral pattern where phloem is on the abaxial side and xylem is on the adaxial side. Within the vascular strands, the xylem and phloem tissues differentiate asymmetrically from the procambium. The patterning of the vascular bundles in the shoot is closely associated with adaxial/abaxial patterning of the lateral organs and with the establishment of central versus peripheral identity within the stem7. Dominant rev-d, phbd or phv-d gain-of-function mutants have adaxialized lateral organs and develop vascular bundles in which xylem surrounds phloem12-14. All of the described Class-III HD-ZIP gain-of-function mutations are located in a microRNA165 (miR165) or miR166 target sequence within a putative sterol /lipid-biding START domain sequence12-13. Mutations that disrupt the miRNA in a similar gain-of-function phenotype, suggests that the HDZIP-III genes are under the control miRNA regulation12,15. YABBY gene family is another gene family responsible for adaxial/abaxial patterning in lateral organs. It expression is located in the abaxial side of the leaves16-17. KANADI gene family members also contribute to abaxial identity in leaves, and are also active in the stem, where YABBY gene family members are not active8. Triple loss-offunction mutants of kan1 kan2 kan3 show radialized amphivasal vascular bundles in stems12. The kan1 kan2 kan3 mutants, phenocopies the rev-d, phb-d or phv-d gain-offunction mutants12, and has abaxial Class III HD-Zip gene expression8. While several plant hormones have been implicated in the regulation of vascular tissue formation, considerable evidence indicates that auxin is the major signal involved in several aspects of the ontogeny of the vascular system20-21. When supported by cytokinins, indole-3-acetic acid (IAA) can induce xylem tracheary element differentiation in suspension culture cells of suitable species11. The findings of Ko and colleges22 after microarrays analyses of secondary growth induced in Arabidopsis plants, suggests that many of the genes up-regulated in wood-forming stems might be regulated by auxin. It is clear that polar auxin transport is crucial for the continuous formation of vascular bundles, however, the precise molecular mechanisms are still mostly unknown23. To understand better these processes, we need to identify more components that are involved in auxin-flow-dependent procambial cell differentiation. Brassinosteroids (BRs) also influence the vascular system. It has been shown that endogenous biosynthesized BRs promote xylem formation and suppress phloem formation24. Late studies have shown that HD-Zip-III genes seem to be regulated by BRs in vascular cells25-26. The vascular differentiation defects seen in BR-deficient mutants form increased amounts of phloem and reduced amounts of xylem27-28. Mutant analysis of the three BR receptors suggests that they act redundantly to regulate vascular differentiation. Possibly signaling through the BR receptors in the procambium induces xylem proliferation and, at the same time, represses phloem proliferation29. Key genetic mechanisms originally characterized as regulating the meristematic cells of the shoot apical meristem are also expressed in vascular cambium during wood growth3. Both KNOX30 and MYB31 transcription factors have been shown to regulate genes that encode key cell wall enzymes. Using xylem and phloem whole transcriptome studies from secondary tissues of Arabidopsis, Zhao et al.32 realized that the same genes involved in the regulation of primary meristems, are also regulators of the cambium. CLAVATA1 and 2 were expressed in the phloem and cambium but not CLAVATA (CLV3). Although, genes belong to the CLE-family, from which CLV3 belongs to, were expressed in this tissues33. 1Berleth, T. 2001. J. Plant Growth Regul. 20:14-21. A. T. 2005. Trends Plant Sci. 10:210-214. 3Groover, A and Robischon, M. 2006. Curr Opin Plant Biol. 9:55-58. 4Ko, J-H., Han, K-H., Park, S. and Yang, J. 2004. Plant Physiol. 135:1069-1083. 5Forest, L., San Martin, J., Padilla, F., Chassat, F., Giroud, F. and Demongeot, J. 2004. Acta Biotheor. 52:415-438. 6Bowman, J.L., Eshed, Y. and Baum, S.F. 2002. Trends Genet. 18:134-141. 7Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F. and Bowman, J.L. 2003. Curr Biol. 14:1768-1774. 8Eshed, Y., Baum, S.F., Perea, J.V. and Bowman, J.L. 2001. Curr. Biol. 11:1251-1260. 9Waites, R. and Hudson, A. 2001. 10Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I. and Morelli, G. 2001. Plant Physiol. 126:643-655. 11Fukuda, H. 1997. Plant Cell. 9:1147-1156. 12McHale, N. and Koning, E. 2004. Plant Cell. 16:1730-1740. 13Juarez, M.T., Kul, J.S., Thomas, J., Heller, B.A. and Timmermans, M.C.P. 2004. Nature. 428:84-88. 14McConnell, J.R., Emery, J.F., Eshed, Y., Bao, N., Bowman, J.L. and Barton, M.K. Nature. 411:1730-1740. 15Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K. and Bartel, D.P. 2005. Development. 128:1923-1932. 16Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, E., Morita, E.H. and Okada, K. 1999. Genes Dev. 13:1079-1088. 17Siegfried, K.R., Eshed, Y., Baum, S.F., Ostuga, D., Drews, G.N. and Bowman, J.L. 1999. Development. 126:4117-4128. 18Bowman, J. 2004. Bioessays. 26:938-942. 19Kerstetter, R.A. Bollman, K. Taylor, R.A., Bomblies, K. and Poething, R.S. 2001. Nature. 411:709-713. 20Aloni, R. 1987. Annu. Rev. Plant Physiol. 38:179-204. 21Sachs, T. 2000. IPlant Cell Physiol. 41:649-656. 22Ko, J.H., Han, K.H. 2004. Plant Mol. Biol. 55:433-453. 23Fukuda, H. 2004. Nat. Rev. Mol. Cell Biol. 5:379-391. 24Nagata, N., Asami, T. and Yoshida, S. 2001. Plant Cell Physiol. 42:1006-1011. 25Ohashi-Ito, K., Demura, T. and Fukuda, H. 2002. Plant Cell Physiol. 43:1146-1153. 26Ohashi-Ito, K. and Fukuda, H. 2003. Plant Cell Physiol. 44:1350-1358. 27Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Kauschmann, A., Altmann, T., Reidei, G.P., Nagy, F., Schell, J. and Koncz, C. 1996. Cell. 85:171-182. 2Groover, 28Choe, S. Noguchi, T., Fujioka, S., Takatsuto, S., Tissie, C. P., Gregory, B. D., et al. 1999. Plant Cell. 11:207-221. 29Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J-C., Nam, H., Li, J. and Chory, J. 2004. Development. 131:5341-5351. 30Mele, G., Sato, N. and Hake, S. 2003. Genes and Development 17:2088-2093. 31Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L., Smith, C., Bevan, M., Mansfield, S., Whetten, R., Sederoff, R. et al. 2003. Plant J. 36:743-754. 32Zhao, C., Craig, J., Petzold, H., Dickerman, A. and Beers, E. 2005. Plant Physiol. 138:803-818. 33Schrader, J., Nilsson, J., Mellerowicz, E., Berlund, A, Nilsson, P., Hertzberg, M. and Sandberg, G. 2004. Plant Cell. 16:2278-2292. TASK Genetic characterization of vascular bundles patterning and development Acting antagonistically to the Class III HD-Zip genes are the KANADI genes, which also encode putative transcription factors. Class III HD-Zip genes are absolutely required for shoot apical meristem formation and have been implicated in functioning in the vascular cambium. While KANADI genes do not appear to be required for proper meristem function, they are needed for pattern formation of organs produced by the shoot apical and vascular meristems. As there are five Class III HD-Zip genes in the Arabidopsis genome the problem of redundancy must be overcome. Our approach to examine Class III HD-Zip gene function in the vasculature relies on the observation that ectopic expression of either KANADI or miR165/166 in the shoot apical meristem leads to loss of Class III HD-Zip gene activity and meristem arrest7. miR165/166 are microRNAs that target Class III HDZip mRNAs for inactivation, perhaps through mRNA cleavage7,15. Based on the work of Ohashi-Ito et al.34 where they used genechip arrays on two Arabidopsis plants expressing ectopically ATHB8 and REV with a mutation in the START domain, and resulting in a higher production of tracheary elements and xylem precursor cells, respectively34, we selected genes involved in cambium and xylem division, late phloem development and early phloem development. In this last group, the gene ALTERED PHLOEM DEVELOPMENT (APL) was the selected, a gene involved in phloem identity in Arabidopsis35. The promoters of the selected genes were already cloned into a plasmid expressing LhG4. These promoters will then drive the expression of REV that is fused with the promoter pOp. REV gene was chosen because is involved in lateral meristem formation, the establishment of lateral organ polarity and xylem differentiation7, 36. The idea is to examine whether we can extend the expression of REV leading to an increased cambial activity as compared to wild-type. Using the LhG4/pOp system of activation, we expressed several CLE-genes under the specific expression of ATHB8 and ATHB15. ATHB8 is induced by auxin and promotes procambial/cambial differentiation into xylem tissues. Its ectopic expression results in promotion of xylem tissue formation. ATHB-15 is also predominantly in vascular tissue. Both genes are regulated by miRNAs. For instant, in the gain-of-function MIR166a mutant, the ATHB-15 transcript level was drastically reduced and the vascular system was severely altered with expanded xylem tissue and interfascicular region, indicating that vascular cell differentiation is greatly promoted37. 34Ohashi-Ito, K, Kubo, M. and Fukuda, H. 2005. Plant Cell Physiol. 46:1646-1656. 35Bonke, M., Thitamadee, S., Mahonen, A.P., Hauser, M.T. and Helariutta, Y. 2003. Nature. 426:181-186. 36Zhong, R. and Ye, Z.H. 2004. Plant Cell Physiol. 45:369-385. 37Kim, J. et al. 2005. Plant J. 42:84-94. Task description A two-component system for trans-activating gene expression will be utilized. In this system, a heterologous transcription factor, GAL4 from yeast can be driven by a variety of promoters, depending on the spatial and temporal expression pattern desired. GAL4 can act on the upstream activating sequence (UAS) that is cloned, along with a minimal promoter, upstream of your gene of interest (in this case, KANADI1 and miR165). As a marker, green fluorescent protein gene (GFP) is also fused to the GAL4 UAS, so that GFP expression correspond to GAL4 expression. Since ectopic expression of either of these transgenes can reduce or eliminate Class III HD-Zip gene expression, loss of Class III HD-Zip gene expression will reflect the expression pattern of the GAL4 driver. We have acquired a number of GAL4 driver lines that represent a diverse set of expression patterns in the roots and hypocotyl (Sabrina Sabatini, personal communication). In this study, lines harboring a transgene in which the upstream activating sequences (UAS) that are cloned along with a minimal promoter upstream of our genes of interest; KANADI1 and miR165, will be crossed to the driver lines to determine whether perturbation in Class III HD-Zip gene activity alters root development, and if so, the nature of the morphological basis of the altered development will be examined. After analyzing the GFP expression and the cross sections of the plants resulting from the crosses between the GAL4 and UAS lines, the ones displaying alterations in the vascular system will be subjected to further genetic analysis. In addition, since auxin is implicated in the establishment of shoot apical meristem and the vascular system, the determination whether auxin acts upstream of Class III HD-ZIP genes in a linear pathway will be determined by crossing these lines to a mutants resistant to the effects of auxin such as axr1 and by adding exogenous auxin to determine if the phenotype is ameliorated. The promoter pOp consist of lac operators cloned upstream of a minimal promoter. Transcription from the promoter is activated by crossing reporter plants with activator lines that expressed a chimeric transcription factor, LhG4. This factor comprises transcription-activation domain-II from GAL4 of Saccharomyces cerevisiae fused to a mutant lac-repressor that binds its operator with increased affinity. The LhG4/pOp system allows coordinating the expression of multiple gene products and restrict transgene phenotypes to the F1 generation. The LhG4 system offers spatially regulated gene expression in the tissues of whole plants growing under normal conditions without the need for external intervention. In other words, it is the possible to use “tissuespecific” promoters to restrict the activity of transgenes to certain tissues when we use a specific promoter driving LhG4. RESULTS Expression of REV gene under the action of promoters of genes involved in the cambium and phloem establishment and development might bring us some insights into vascular development, specifically whether manipulation of its expression could lead to continued cambial activity. Also, we have expressed a number of CLE family members under two promoters, ATHB8 and ATHB15, expressed in vascular tissue10, 31. These genes can may play a role in the cambium as they were detected in the cambial meristem array performed by Schrader et al.33. In addition, one of the GAL4 lines when crossed with UAS::miR165, resembles bri1 mutants38, with rosette leaves curly and round. Another line has a thick stem in comparison with wild type plants after induction of secondary growth. These will be compared to control lines in which GAL4 lines are crossed to Arabidopsis thaliana ecotype Ler. 38Wang, Z et al., J. 2001. Nature. 410:380-383. REPERCUSSIONS Most Class III HD-Zip genes appear to regulate vascular development in a redundant in manner. However, the general roles for Class III HD-Zip genes in vascular development are still unknown. Other factors such as phytohormones, such as brassinosteroids, are also involved in vascular differentiation and may act in combination with Class III HD-Zip genes. For example, early physiological studies using Zinnia explants have shown that BRs play an important role in promoting xylem differentiation. The levels of BR intermediates have been shown to peak at the transition from undifferentiated cells to tracheary elements. In Arabidospsis two mutants defective in BR-synthesis, cpd and dwarf7, exhibit vascular differentiation effects27, 28. However, vascular phenotypes in BR biosynthetic and signaling mutants have not been characterized in detail. Therefore, we expect to elucidate the important role of Class III HD-Zip and KANADI genes, BR and eventually, auxin during the establishment and development of the vascular system. Arabidopsis is the species chosen for this study because it offers all the genetic tools associated with it. This way, and given the high degrees of anatomical and genomic similarities regarding wood formation in Arabidopsis compared with that in poplar39 and loblolly pine40 discoveries connected with these transcript profiles are also likely to lead to important applications in economically important tree species. We drove our attention into secondary growth, or in other words, wood formation, due to the economical importance throughout the world and above all, the imperative need for maintenance of the ecosystem balance. We believe that the contribution of this work into the scientific world would be important but most importantly, for the possible impact in wood production and the consequent management of the territory, especially in Portugal. For instance, for the last 10 years, one fifth of the Portuguese forest has disappeared due to fires. The possibility of having trees producing more wood in shorter period of time and occupying a reduced area of land would allow a more efficient reforestation and would have an impact in the economy of the more rural areas of the country. 39Chaffey, N., Cholewa, E., Regan, S. and Sundberg, B. 2002. Physiol Plant. 114:594600. 40Kirst, M., Johnson, A., Baucom, C., Ulrich, E., Hubbard, K., Staggs, R., Paule, C., Retzel, E., Whetten R. and Sederoff, R. 2003. Proc. Natl. Acad. Sci. USA 100:7383-7388.