pmic7095-sup-0004-s2
advertisement

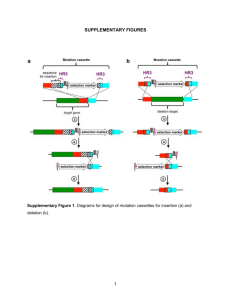
Supplementary Method for Proteomic analysis of spontaneous mutants of Lactococcus lactis: involvement of GAPDH and arginine deiminase pathway in H2O2 resistance. Tatiana Rochat(+)*, Samira Boudebbouze, Jean-Jacques Gratadoux, Sébastien Blugeon, Philippe Gaudu, Philippe Langella and Emmanuelle Maguin(+) INRA, Institut Micalis, UMR1319/INRA-AgroParisTech, Jouy-en-Josas F78350, France. * present address: Institut de Génétique et Microbiologie, CNRS, UMR 8621, IFR115, Centre Scientifique d'Orsay, Université Paris Sud 11, Orsay, France. + Corresponding authors: T. Rochat, Institut de Génétique et Microbiologie, Université Paris-Sud Bât 400, 91405 Orsay Cedex (FR); tatiana.rochat@u-psud.fr; Phone: +33(0)169154655; Fax: +33(0)169156678. E. Maguin, INRA, Institut Micalis, Bât 440, Domaine de Vilvert, 78352 Jouy-en-Josas cedex (FR); emmanuelle.maguin@jouy.inra.fr; Phone: +33(0)134652518; Fax: +33(0)134652727. Inactivation of gapA, arcA and arcB in L. lactis. To disrupt genes in L. lactis strains, we used pRV300 vector which is composed of a pBluescript SKreplicon for propagation in E. coli and an erythromycin resistance marker. An internal fragment of the gene to disrupt was inserted into pRV300 and the resulting plasmids were able to integrate into the chromosome by homologous recombination as single copies and were maintained stably [1]. After chromosomal integration of pRV300, the targeted gene was disrupted while the production of a Cterminal truncated form of the protein could remain. Two pRV300:gapA plasmids were constructed for gapA disruption: the chromosomal integration led to a protein truncated after the 219th amino-acid for the first one (pRV300:gapA*) and after the 148th amino-acid for the second one (pRV300:gapA**). Consequently, the gapA mutant resulting from pRV300:gapA* integration could produce a truncated form of GapA which contains Cys152 residue contrary the one resulting from pRV300:gapA**. Internal fragments of gapA were amplified by PCR using primers designed from the database sequence of L. lactis MG1363 [2]: 1) for gapA* fragment with gapA5' (5'- ggtcaaagaagatggttttgatgtcaacgg) and gapA3' (5'-gctggaacaatattttcagctgcagcacgc); 2) for gapA** fragment with gapAs5' (5'-GATCCTGCAGGGTAGTTAAAGTTGGTATTAACGG) and gapAs3' (5'GATCAAGCTTATTAAGAAATAACTGTTTCACTTCC). The PCR products (named gapA* and gapA**) were inserted in the cloning vector pCRII-TOPO (Invitrogen) and sequenced. The gapA* and gapA** fragments were then inserted into pRV300 after digestion, resulting in pRV300:gapA* and pRV300:gapA** which were transformed and subsequently purified from E. coli Top10 strain (Invitrogen). pRV300:gapA* and pRV300:gapA** were then electroporated in J60011, SpOx2 and SpOx3 mutants. To disrupt arcA and arcB genes in SpOx1 mutant, internal fragments of arcA or arcB genes of 577 or 525 bp respectively, were amplified by PCR using primers: arcA5’ (5’- gcgaattcggacgtacttatgatggattgactg) and arcA3’ (5’-ttgcggccgcccataattcctgggaaaactgtaaattg) or arcB5’ (5’-gcgaattcgaccttggggctcatcctga) and arcB3’ (5’-ttgcggccgccccagttagattctcccattgataccc). The PCR products (named arc*) were inserted into pRV300 after digestion using EcoRI and NotI, resulting in two pRV300:arc* plasmids which were transformed and purified from E. coli. After sequencing of the cloned arc* fragments, pRV300:arc* plasmids were electroporated into SpOx1 strain, resulting in two SpOx1 arcA and arcB mutants. Bacterial strains and plasmids used in this study. Strains Characteristics References MG1363 plasmid-cured L. lactis ssp cremoris used as wild type strain [3] J60011 parental strain of SpOx mutants; lac+/prt+ MG1363 derivative generated by conjugation with NCDO 712, containing the pLP712 conjugative plasmid [4] SpOx1 spontaneous mutant of J60011, H2O2 resistant [4] SpOx2 spontaneous mutant of J60011, H2O2 resistant [4] SpOx3 spontaneous mutant of J60011, H2O2 resistant [4] VEL11998 gapA mutant of J60011 (Emr) , pRV300:gapA* integration This work VEL12074 gapA mutant of L. lactis SpOx2 (Emr), pRV300:gapA* integration This work VEL11999 gapA mutant of L. lactis SpOx3 (Emr), pRV300:gapA* integration This work VEL12393 gapA mutant of L. lactis J60011 (Emr), pRV300:gapA** integration This work VEL12394 gapA mutant of L. lactis SpOx2 (Emr), pRV300:gapA** integration This work VEL12395 gapA mutant of L. lactis SpOx3 (Emr), pRV300:gapA** integration This work VEL12278 arcA mutant of L. lactis SpOx1 (Emr) This work VEL12280 arcB mutant of L. lactis SpOx1 (Emr) This work NZ9000 Plasmids MG1363 carrying nisRK genes on the chromosome Characteristics [5] References pRV300 Apr/Emr carrying 453 bp internal fragment of gapA This work pRV300:gapA** pRV300 Apr/Emr carrying 444 bp internal fragment of gapA This work pRV300:arcA* pRV300 Apr/Emr carrying 577 bp internal fragment of arcA This work pRV300:arcB* pRV300 Apr/Emr carrying 525 bp internal fragment of arcB This work pRV300:gapA* pRV300 pAM1, Emr [1] pVE3655 pWV01, Cmr carrying PnisA, a nisin-inducible promoter [6] pNZ:gapA pWV01, Cmr carrying gapA under control of PnisA [2] Apr, Emr and Cmr: resistance to ampicillin, erythromycin and chloramphenicol, respectively. [1] Leloup, L., Ehrlich, S. D., Zagorec, M., Morel-Deville, F., Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol 1997, 63, 2117-2123. [2] Willemoes, M., Kilstrup, M., Roepstorff, P., Hammer, K., Proteome analysis of a Lactococcus lactis strain overexpressing gapA suggests that the gene product is an auxiliary glyceraldehyde 3phosphate dehydrogenase. Proteomics 2002, 2, 1041-1046. [3] Gasson, M. J., Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 1983, 154, 1-9. [4] Rochat, T., Gratadoux, J. J., Corthier, G., Coqueran, B., et al., Lactococcus lactis SpOx spontaneous mutants: a family of oxidative-stress-resistant dairy strains. Appl Environ Microbiol 2005, 71, 27822788. [5] Kuipers, O. P., P. G. de Ruyters, M. Kleerezen, Vos, W. M. d., Quorum sensing-controlled gene expression in lactic acid bacteria. J.Biotechnol. 1998, 64, 15-21. [6] Le Loir, Y., Nouaille, S., Commissaire, J., Bretigny, L., et al., Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microbiol 2001, 67, 4119-4127.