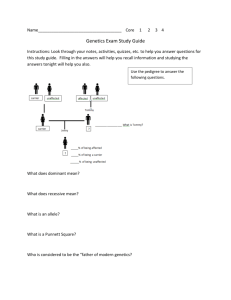
Figure 1. Distribution and levels of tTA mRNA and protein A
advertisement

Figure 1. Distribution and levels of tTA mRNA and protein A. Northern blot including olfactory epithelium (OE), spinal cord (SC) and brain (B) extracts, showing two bands with the tTA probe in double transgenic mice. From a single representative double transgenic line (997 EGFP-TRE-hG93ASOD1cDNA). B. Western blot of protein distribution across areas in A (anti-tTA monoclonal; MobiTec, Göttingen, Germany). Positive control: homogenates from the activator PrP-tTA 959 mouse line. From the same representative transgenic line. C. Western blot for the nuclear localization of the tTA protein across the nervous system areas. From the same representative transgenic line. Numbers indicate protein molecular weights. D. Quantitative analysis of EGFP and human SOD1 production related to tTA levels. To determine if the production of responder elements was dose-dependent on tTA, activator and responder homozygous genotypes were established for the 997 EGFP-TRE-hG93ASOD1cDNA line. To obtain homozygous genotypes, double transgenic mice were crossed to activator PrP-tTA mice or the corresponding responder line. Hemizygous littermates were used for comparison. The genotypes were established by real-time PCR. Data are all from groups of 2-3 mice, and experiments were replicated twice. The densiometric analysis of bands was performed with a computer-assisted image analysis system (AIS 3.0-Imaging Research) All samples were normalized to -actin levels. E. Northen and Western blots of representative samples quantified in D. The responder EGFP and human SOD1 mRNA and/ or protein levels are irrespective of tTA levels (observe right panel in E). This suggestes that non-limiting theresholds caused the transcriptional impairments observed. AA, homozygous activator genotypes; RR, homozygous responder gentotypes; Aa, hemizygous activator genotypes; Rr, hemizygous responder genotypes; 1, littermates harbouring 25% to 50% (poor) FVB/N strain content; 2, littermates harbouring 50% to 75% (enriched) FVB/N content; 3; double transgenic mice with 50% FVB/N content, relative to the set of crosses to obtain the homozygous genotypes. Name F.SOD2 R.SOD2 F.EGFP R.EGFP F.SOD1 R.SOD1 F.tTA R.tTA Primers for screenings and analysis of cDNA expression Sequence Anealing temperature and size amplicon 5’-TGCACTGAAGTTCAATGGTGG-3’ 56 °C; 238 bp 5’-TAGAGCAGGCAGCAATCTGT-3’ 5’-ACCACATGAAGCAGCACGACT-3’ 60 °C; 490 bp 5’-CTTGTACAGCTCGTCCATGCC-3’ 5’-GATTCCATGTTCATGAGTTTGG-3’ 56 °C; 360 bp 5’-GGCCTCAGACTACATCCAAG-3’ 5’-GAGCAGCCTACATTGTATTGG-3’ 56 °C; 458 bp 5’-GGCCGAATAAGAAGGCTGG-3’ F-PCMV1 R-PCMV1 Primers for real-time PCR analyses 5’-AGGCGTGTACGGTGGGAGG-3’ 60 °C; 150 bp 5’-CCCGGGTACCGAGCTCGAA-3’ F xist R xist 5’-TGATACGGCTATTCTCGAGCC-3’ 5’-TCAAGGCGAATCCCGCAACC-3’ F QtTA R QtTA 5’-AGAGCAGCCTACATTGTATTGG-3’ 60°C; 99 5’-AAGGGCAAAAGTGAGTATGGTG-3’ control F.tetSOD1 60 °C; 137 bp, GeneBank NT_039711. Normalisating purposes. bp. Internal Primers for cloning 5’-CTGTGCAGTCCTCGGAACCAGGA-3’ R.tetSOD1 5’-AAGGAAAGAAGCTAGCAGGATAACAGATGAGTTAAGGG-3’ Table 1. Primer sequences.