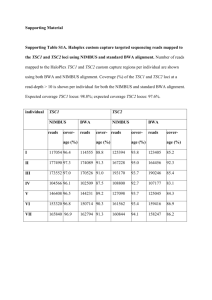
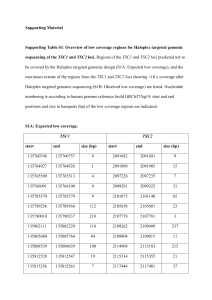
Supplemental Materials and Methods Mouse Studies. The MMTV
advertisement

SUPPLEMENTAL MATERIALS AND METHODS Mouse Studies. The MMTV-PyMT allele was bred onto the C57BL/6 background for at least seven generations. Tsc2+/- female mice (C57Bl/6 background) were crossed with MMTV-PyMT male mice and pups genotyped using primers that distinguish between wild-type (wt) and mutant Tsc2 alleles 5’ CAAACCCACCTCCTCAAGCTTC3’; (H162: H163: 5’ AATGCGGCCTCAACAATCG3’; H164: 5’AGACTGCCTTGGGAAAAGCG3’) and the PyMT transgene (Forward: 5’ GGAAGCAAGTACTTCACAAGGG3’; Reverse: 5’ GGAAAGTCACTAGGAGCAGGG3’). PyMT/Tsc2+/+ and PyMT/Tsc2+/- female mice were palpated twice weekly beginning at 6 weeks of age to monitor tumor onset, and was continued for 8 weeks thereafter. Mice were sacrificed when tumors had reached a maximum of 2 cm3 for isolated tumors or 6 cm3 in total for all tumors. For histological analysis, mammary and lung tissues were excised and stored in 10% neutral buffered formalin for 48 hours before embedding in paraffin. Mammary tumors were sectioned at 5 µm and lung tissues sectioned in steps at 50 µm. For treatment studies, mice were injected intra-peritoneal daily with either RAP (8 mg/kg) or vehicle (5.2% PEG 400 and 5.2% Tween-20) for 14 consecutive days prior to sacrifice. RNAi and Retroviral Transduction. MicroRNA-based shRNAs (shRNAmir) targeting eIF4E (sh4E.389 and sh4E.610) or the neutral control Firefly Luciferase (shFLuc.1309) (Premsrirut et al 2011) cloned in retroviral expression vectors were used in these studies. Virus infections (performed by spinoculation of 2x105 target cells) were repeated every 12 h for a total of 3 cycles. Infected cells were selected 5 days with either hygromycin (2.5 mg/ml) for DOXinducible vectors or puromycin (3 mg/ml) for MLP-based vectors. For transient knockdown, MDA-MB-231 cells were transfected with either non-targeting (NT) siRNA or siRNAs targeting human eIF4E (Dharmacon) using lipofectamine 2000 according to the manufacturer’s recommendations (Invitrogen). Antibodies used for Western blot analysis were directed against eIF4E (Santa Cruz #sc9976), VEGF (Santa Cruz #sc507), MMP9 (Santa Cruz #sc6840), Cyclin D1 (cell signaling#2926), β-actin (Sigma#A5316), and α-tubulin (Sigma #T5268). The relative intensity of signals from immunoblots were quantitated using ImageJ program. Primers used for Quantitative RT-PCR. Primers used for qRT-PCR (Fig. 6) were: MMP9f ( 5’ TCCAACTCACTCACTGTGGTTGCT3’) and MMP9r ( 5’ AGACTGCCAGGAAGACACTTGGTT3’), 5’ TGCCAGCAACATTACCACAGTGTC3’) 5’ TCTTCATCCAGCTCCTTGTTGGGT3’), VEGFCf and ( VEGFCr Cyclin ( D1f (5’CAGGTTCCTGTTCACAATACCTCA3’) and Cyclin D1r (5’AGACCGCCCACCTGCC3’), and GAPDHf (5’GAAGGTCGGTGTGAACGGATTTGGC3’) and (5’GATGGGCTTCCCGTTGATGACAAGC3’). Error bars represent the SEM. GAPDHr SUPPLEMENTAL FIGURE LEGENDS Figure S1. Changes in tumor size as a function of time post-onset. n=12 for each genotype. Error bars denote the standard deviation. Figure S2. Quantitation of TSC2 and p-S6 levels in primary tumors from mice of the indicated genotypes. Sections from tumor samples were processed for immunohistochemical staining (see Fig. 1C) and the intensity of the staining was quantitated using Aperio ImageScope. Values represent the average of 5 fields from 3 different sections. The relative intensity of staining is denoted as 0, 1+, or 2+, with 2+ indicating the most intense signal. Figure S3. Genotyping of normal tissues and tumors from PyMT/Tsc2+/+ and PyMT/Tsc2+/mice. Semi-quantitative PCR was used to genotype the Tsc2 locus in DNA from normal tissue of PyMT/Tsc2+/- (lane 2) or PyMT/Tsc2+/+ (lanes 3 and 4) mice or from breast tumors obtained from PyMT/Tsc2+/- mice (lanes 5-9). The position of migration of the mutant and wild-type alleles are indicated to the right. M denotes molecular weight markers (lane 1). Figure S4. Quantitation of TSC2 and p-S6 levels present in pulmonary metastasis from mice of the indicated genotypes treated with vehicle or rapamycin. Sections from lungs were processed for immunohistochemical staining (see Fig. 2B) and the intensity of the staining was quantitated using Aperio ImageScope. Values represent the average of 5 fields from 3 different sections. The relative intensity of staining is denoted as 0, 1+, 2+, or 3+ with 3+ indicating the most intense signal. Figure S5. Representative TUNEL assay and Ki-67 staining of mammary tumors from PyMT/Tsc2+/+ and PyMT/Tsc2+/- mice treated with rapamycin or vehicle. Quantitation of data is presented in Figures 2D and 2E. Bar, 50 µm. Figure S6. A. 35S-Methionine metabolic labelling of TM15 cells pre-treated with the indicated concentrations of rapamycin, silvestrol, or hippuristanol. Cells were exposed to compounds for two hours in methionine-free medium after which 35S-methionine (150-220 µCi/ml) was added for 15 minutes. Cells were washed with cold PBS, and lysed in RIPA buffer (50 mM TrisHCl7.5, 150 mM NaCl, 1 mM DTT, 0.1% SDS, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1 mM Phenylmethylsulfonyl fluoride (PMSF), 1 µg/ml each of leupeptin, pepstatin, and aprotinin). The amount of trichloroacetic acid-insoluble [35S]-labelled protein was quantitated by liquid scintillation counting. Incorporation was set relative to vehicle-treated TM15 cells. N=3; error bars represent the SEM. B. Fraction of viable TM15 cells upon exposure to rapamcin, silvestrol, or hippuristanol. TM15 cells (2.5 x 104) were cultured in 96-well plates and treated with the indicated concentrations of rapamycin, silvestrol, and hippuristanol for 24 hours. Cells were fixed with 50% (w/v) trichloroacetic acid for one hour and stained with 0.4% SRB (sulforhodamine B in 1% acetic acid) for 10 min, after which excess dye was removed by repeated washing with 1% (v/v) acetic acid. The dye was solubilised in 10 mM Tris base and quantitated by absorbance at OD490. N=3; error bars represent the SEM. Figure S7. A. Immunoblot analysis of MLP/sh4E.610- and shFLuc.1309-infected TM15 cells. Extracts were prepared for blotting after selection of infected cells with puromycin. The quantitation for relative eIF4E levels is provided below the blot and is from 3 independent experiments. B. Representative crystal violet staining of migrating and invading MLP/sh4E.610or shFLuc.1309-infected TM15 cells. C. Quantitation of cell migration (left panel) and matrigel invasion (right panel) of MLP/sh4E.610- and shFLuc.1309-infected TM15 cells. Data representing three different experiments with five fields counted per experiment. Bars represent SEM. *, p=0.007. Figure S8. Quantitation of eIF4E and GFP levels present in primary tumors and pulmonary metastasis from allografts derived from TM15 cells expressing the indicated shRNAs. Tissue sections were processed for immunohistochemical staining (see Fig. 4C) and the intensity of the staining was quantitated using Aperio ImageScope. Values represent the average of 5 fields from 3 different sections. The relative intensity of staining is denoted as 0, 1+, 2+, or 3+ with 3+ indicating the most intense signal. REFERENCES Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C et al (2011). A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 145: 145-158.